Abstract
Pleiotrophin is a pleiotropic cytokine that has been demonstrated to have a critical role in regulating energy metabolism, lipid turnover and plasticity of adipose tissue. Here, we hypothesize that this cytokine can be involved in regulatory processes of glucose and lipid homeostasis in the liver during pregnancy. Using 18‐days pregnant Ptn‐deficient mice, we evaluated the biochemical profile (circulating variables), tissue mRNA expression (qPCR) and protein levels of key enzymes and transcription factors involved in main metabolic pathways. Ptn deletion was associated with a reduction in body weight gain, hyperglycemia and glucose intolerance. Moreover, we observed an impairment in glucose synthesis and degradation during late pregnancy in Ptn −/− mice. Hepatic lipid content was significantly lower (73.6%) in Ptn −/− mice and was associated with a clear reduction in fatty acid, triacylglycerides and cholesterol synthesis. Ptn deletion was accompanying with a diabetogenic state in the mother and a decreased expression of key proteins involved in glucose and lipid uptake and metabolism. Moreover, Ptn −/− pregnant mice have a decreased expression of transcription factors, such as PPAR‐α, regulating lipid uptake and glucose and lipid utilization. Furthermore, the augmented expression and nuclear translocation of glycerol kinase, and the decrease in NUR77 protein levels in the knock‐out animals can further explain the alterations observed in hepatic glucose metabolism. Our results point out for the first time that pleiotrophin is an important player in maintaining hepatic metabolic homeostasis during late gestation, and further highlighted the moonlighting role of glycerol kinase in the regulation of maternal glucose homeostasis during pregnancy.
Keywords: glycerol kinase, liver, metabolism, NR4A1, pleiotrophin, pregnancy
Abbreviations
- ACC
acetyl‐CoA carboxylase
- AUC
area under the curve
- GIP
glucose‐induced insulinotropic peptide
- GTT
glucose tolerance test
- Gyk
glycerol kinase gene
- IDL
intermediate density lipoproteins
- LDL
low density lipoproteins
- NEFA
non‐esterified fatty acids
- NUR77
mice isoform for NR4A1
- PTN
pleiotrophin
- Ptn
pleiotrophin gene
- TBP
TATA‐binding protein
- VLDL
very low density lipoproteins
1. INTRODUCTION
During pregnancy, the mother experiences significant metabolic, anatomical and physiological changes. 1 , 2 In the first two‐thirds of pregnancy, the mother is in an anabolic stage characterized by hyperphagia, increased insulin sensitivity 3 and the store of lipids in the adipose tissue. 4 These fat depots are able to account for up to 28% of maternal weight both in women 5 and in rats. 6 , 7
On the contrary, the last third of gestation is characterized by a catabolic state, 2 , 8 hyperinsulinemia, reduced insulin sensitivity, and enhanced placental transference of nutrients. Maternal glucose is intensively transferred to the fetal site that leads to maternal hypoglycemia, despite the increased rate in maternal gluconeogenesis. In parallel, maternal tissues adapt their metabolism to use alternative energetic substrates such as fatty acids and ketone bodies. Accordingly, lipolysis of triacylglycerides is enhanced in maternal adipose tissue and consequently, free fatty acids and glycerol are released from adipose tissue. 3 Free fatty acids are used for β‐oxidation and ketone bodies synthesis, and glycerol is used for hepatic gluconeogenesis. In fact, glycerol is preferred to amino acids for hepatic de novo gluconeogenesis during late pregnancy when compared to a non‐pregnant state. 9 Additionally, lipid turnover is increased during late pregnancy. 10 The fatty acids released by the enhanced lipolysis in the adipose tissue can be re‐esterified in the liver increasing very low density lipoproteins (VLDL) production and leading to hypertriglyceridemia in maternal plasma. 2
Pleiotrophin (PTN) or heparin‐binding growth‐associated molecule (HB‐GAM) is a highly conserved cytokine. 11 PTN is expressed in cells, in early differentiation stages and especially during embryonic development. 12 The pleiotropic functions of PTN include mitogenicity, angiogenesis, inflammation, cell differentiation and mechanisms of the stem cell renewal. 13 In the liver, PTN has been shown to act as a mitogen on hepatocytes in developmental and pathological processes 14 , 15 such as liver fibrosis or hepatitis. 12 , 16 , 17 Even though research has been carried out to understand the role of PTN in liver pathologies, little is known about its involvement in hepatic metabolism during physiological conditions associated with metabolic adaptations, as pregnancy. Thus, the aim of this study is to determine the role of pleiotrophin in glucose/insulin homeostasis and liver metabolism during pregnancy.
2. MATERIALS AND METHODS
2.1. Animals
PTN genetically deficient (Ptn −/−) mice were a kind gift from T. F. Deuel (The Scripps Research Institute, La Jolla, CA, USA). 18 , 19 Female Ptn+/+ (n = 15) and Ptn −/− (n = 19) mice were housed in controlled conditions at 20–22°C with 12 h light/dark cycles, and free access to water and a chow diet (Panlab, Barcelona, Spain). 8–10 weeks old female mice were mated, and after verifying the presence of a vaginal plug, that day was considered as day 0 of gestation (P0). On day 18 (P18), one first group of Ptn +/+ (n = 10) and Ptn −/− (n = 10) mice were fasted for 6 h, exposed to carbon dioxide and killed by decapitation. Plasma and tissues were collected and preserved at −80°C. Animals were maintained in accordance with European Union Laboratory Animal Care Rules (86/609/ECC directive) and protocols were approved by the Animal Research Committee of CEU San Pablo University.
2.2. Plasma analysis, insulin determination and glucose tolerance tests
Plasma insulin levels were determined using immunoassay kits (Mercodia, Sweden). Enzymatic colorimetric tests were used for the determination of glucose (glucose oxidase‐peroxidase method; Spinreact, Spain), triacylglycerols (lipoprotein lipase [LPL]/glycerol kinase [GK]/glycerol phosphate oxidase [GPO]/peroxidase [PDO], Spinreact, Spain), Non esterified fatty acids (NEFA) (Acyl‐CoA synthase –Acyl‐CoA oxidase [ACS‐ACOD] method; Wako Chemicals, Germany), cholesterol (cholesterol esterase [CHE]/cholesterol oxidase [CHOD]/peroxidase [POD]; Spinreact, Spain) and glycerol (Free glycerol reagent kit; Sigma‐Aldrich, USA). Plasma lipoprotein profiles were analyzed by agarose electrophoresis (SAS‐MX Lipo Kit, Helena Biosciences, UK). Plasma glucagon and GIP were measured with a Bioplex pro‐Mouse diabetes‐Immunoassay kit (Bio‐Rad, USA).
Glucose tolerance tests (GTTs) were performed in a second group of 6 h fasted animals (Ptn+/+ (n = 5) and Ptn −/− (n = 9)) using intraperitoneal injection of glucose (2 g/kg). AUCs (area under the curve) for glucose were calculated.
2.3. Hepatic lipid content
Hepatic lipids were extracted in chloroform‐methanol 3:1 and subsequently purified. 20 The obtained different lipid fractions were resolved by thin‐layer chromatography and quantified. 21
2.4. Quantitative real‐time PCR in liver tissue
Total RNA Isolation Kit (Nzytech, Portugal) was used to isolate RNA that was retrotranscribed using the first‐strand cDNA Synthesis Kit (Nzytech, Portugal). Quantitative real‐time PCR (qPCR) analysis was performed using the SYBR green method (Quantimix Easy Kit, Biotools, Spain); the primer sequences are shown in Table 1. The dynamic range for each primer pair was assessed with a standard curve using a pooled sample and the appropriate dilution factors. Amplification efficiency for all primer pairs was determined from the slope of the standard curve and was in the range between 90% and 110%. The relative expression of each mRNA was normalized against Rpl13, used as a reference gene.
TABLE 1.
Primer sets used for qPCR analysis
| Gene | Primer forward | Primer reverse |
|---|---|---|
| Acadl | 5′‐CTTGGGAAGAGCAAGCGTACT‐3′ | 5′‐CTGTTCTTTTGTGCCGTAATTCG‐3′ |
| Acadvl | 5′‐GGAGGACGACACTTTGCAGG‐3′ | 5′‐AGCGAGCATACTGGGTATTAGA‐3′ |
| Acat2 | 5′‐CACGAAATGGGGACAACTG‐3′ | 5′‐AACCTGCCGTCAAGACA‐3′ |
| Acc | 5′‐GTCCCCAGGGATGAACCAATA‐3′ | 5′‐GCCATGCTCAACCAAAGTAGC‐3′ |
| Acox1 | 5′‐TCGAAGCCAGCGTTACGAG‐3′ | 5′‐ATCTCCGTCTGGGCGTAGG‐3′ |
| ApoB100 | 5′‐AAGCACCTCCGAAAGTACGTG‐3′ | 5′‐CTCCAGCTCTACCTTACAGTTGA‐3′ |
| ApoC2 | 5′‐GCATGGATGAGAAACTCAGGG‐3′ | 5′‐AAAATGCCTGCGTAAGTGCTC‐3′ |
| Aqp9 | 5′‐CTATGACGGACTCATGGCCTTT‐3′ | 5′‐ATGAACGCCGTTCCATTTTCT‐3′ |
| Acly | 5′‐AAGCCTTTGACAGCGGCATCATTC‐3′ | 5′‐TTGAGGATCTGCACTCGCATGTCT‐3′ |
| Cebpa | 5′‐GGAACTTGAAGCACAATCGATC‐3′ | 5′‐TGGTTTAGCATAGACGTGCACA‐3′ |
| Cpt1α | 5′‐ACCCTGAGGCATCTATTGACAG‐3′ | 5′‐ATGACATACTCCCACAGATGGC‐3′ |
| Dgat 1 | 5′‐GCCCCATGCGTGATTATTGC‐3′ | 5′‐CACTGGAGTGATAGACTCAACCA‐3′ |
| Dgat 2 | 5′‐AACCGAGACACCATAGACTACTT‐3′ | 5′‐CTTCAGGGTGACTGCGTTCTT‐3′ |
| Fatp4 | 5′‐CCAGTAGTGTGGCCAACTTCCT‐3′ | 5′‐CCACAGACCCACAAACTCATTG‐3′ |
| Fatp5 | 5′‐GAATCGGGAGGCAGAGAACT‐3′ | 5′‐AGCGGGTCATACAAGTGAGC‐3′ |
| Fas | 5′‐AGAGATCCCGAGACGCTTCT‐3′ | 5′‐GCCTGGTAGGCATTCTGTAGT‐3′ |
| Fbp1 | 5′‐CAGGGACGTGAAGATGAAGAAGAA‐3′ | 5′‐TTGTTGGCGGGGTATAAAAAGA‐3′ |
| Fgf21 | 5′‐GGAGCTCTCTATGGATCGCCT‐3′ | 5′‐TGTAACCGTCCTCCAGCAGC‐3′ |
| Gck | 5′‐TAAAGATGTTGCCCACCTACG‐3′ | 5′‐GGAATACATCTGGTGTTTCGTCT‐3′ |
| G6pc | 5′‐CGACTCGCTATCTCCAAGTGA‐3′ | 5′‐GTTGAACCAGTCTCCGACCA‐3′ |
| G6pd | 5′‐CTGGAACCGCATCATAGTGGAG‐3′ | 5′‐CCTGATGATCCCAAATTCATCAAAATAG‐3′ |
| Glut2 | 5′‐TCCCTTGGTTCATGGTTGCT‐3′ | 5′‐GGAAGTCCGCAATGTACTGGA‐3′ |
| Gpat | 5′‐ACGCACACAAGGCACAGAG‐3′ | 5′‐TGCTGCTCAGTACATTCTCAGTA‐3′ |
| Gyk | 5′‐GTCAGCAACCAGAGGGAAACC‐3′ | 5′‐CCACGGCATTATAGAGAGGCT‐3′ |
| Hadha | 5′‐AGCAACACGAATATCACAGGAAG‐3′ | 5′‐AGGCACACCCACCATTTTGG‐3′ |
| Hmgcr | 5′‐TGACCTTTCTAGAGCGAGTGC‐3′ | 5′‐GTGCCAACTCCAATCACAAG‐3′ |
| Hmgcs | 5′‐CGGATCGTGAAGACATCAACTC‐3′ | 5′‐CGCCCAATGCAATCATAGGAA‐3′ |
| Ldh | 5′‐TGTCTCCAGCAAAGACTACTGT‐3′ | 5′‐GACTGTACTTGACAATGTTGGGA‐3′ |
| Lipc | 5′‐TTCTCGGAGCAAAGTTCACCTAAT‐3′ | 5′‐GTGATTCTTCCAATCTTGTTCTTC‐3′ |
| Lpin2 | 5′‐AGTTGACCCCATCACCGTAG‐3′ | 5′‐CCCAAAGCATCAGACTTGGT‐3′ |
| Lpl | 5′‐TGGAGAAGCCATCCGTGTG‐3′ | 5′‐TCATGCGAGCACTTCACCAG‐3′ |
| Mdk | 5′‐TGATGGGAGCACTGGCAC‐3′ | 5′‐CATTGTACCGCGCCTTCTT‐3′ |
| Nur77 | 5′‐TTGAGTTCGGCAAGCCTACC‐3′ | 5′‐GTGTACCCGTCCATGAAGGTG‐3′ |
| Pc | 5′‐GGCGCATGAGTTCTCCAACA‐3′ | 5′‐GTAGGCCCGGGTGTAATTCTC‐3′ |
| Pdha1 | 5′‐GAAATGTGACCTTCATCGGCT‐3′ | 5′‐TGATCCGCCTTTAGCTCCATC‐3′ |
| Pepck | 5′‐AAGCATTCAACGCCAGGTTC‐3′ | 5′‐GGGCGAGTCTGTCAGTTCAAT‐3′ |
| Pfk1 | 5′‐GAACTACGCACACTTGACCAT‐3′ | 5′‐CTCCAAAACAAAGGTCCTCTGG‐3′ |
| Pgc1α | 5′‐CCCTTCTTTGCCATTGAATC‐3′ | 5′‐AATGTTAGGAAAGTTTAGCATCTGG‐3′ |
| Pk | 5′‐CTGGATGGGGCTGACTGTAT‐3′ | 5′‐GGCGTAGCTCCTCAAACAAC‐3′ |
| Plin2 | 5′‐GACAGGATGGAGGAAAGACTGC‐3′ | 5′‐GGTAGTCGTCACCACATCCTTC‐3′ |
| Pparα | 5′‐CACGCATGTGAAGGCTGTAA‐3′ | 5′‐CAGCTCCGATCACACTTGTC‐3′ |
| Pparβδ | 5′‐ACCCTTTGTCATCCACGA‐3′ | 5′‐GGATGTTCTTGGCGAACT‐3′ |
| Pparγ1 | 5′‐TTTAAAAACAAGACTACCCTTTACTGAAATT‐3′ | 5′‐AGAGGTCCACAGAGCTGATTCC‐3′ |
| Rpl13 | 5′‐GGTGCCCTACAGTTAGATACCAC‐3′ | 5′‐TTTGTTTCGCCTCCTTGGGTC‐3′ |
| Rxrα | 5′‐ACATCTGCGCTATCTGTGGG‐3′ | 5′‐TGTCGATCAGGCAGTCCTTG‐3′ |
| Scd1 | 5′‐TTCTTGCGATACACTCTGGTGC‐3′ | 5′‐CGGGATTGAATGTTCTTGTCGT‐3′ |
| Soat2 | 5′‐AGACTTGGTGCAATGGACTCGAC‐3′ | 5′‐CATAGGGCCCGATCCAACAG‐3′ |
| Srebp1 | 5′‐AGGCCATCGACTACATCCG‐3′ | 5′‐TCCATAGACACATCTGTGCCTC‐3′ |
2.5. Protein extraction and western‐blot analysis
Liver tissue was homogenized using a buffer containing 30 mM HEPES, 5 mM EDTA, 1% Triton X‐100, 0.5% sodium deoxycholate and 2 mM of protease inhibitor mixture Pefabloc (Roche Diagnostics, Spain). Protein concentration was measured by the Pierce BCA protein assay kit (Thermo‐Scientific, Rockford, USA).
For nuclear extraction, liver tissue was homogenized in saline buffer and after centrifugation, the pellet was resuspended in a cold buffer A (10 mM HEPES, 0.2 mM EDTA, 10 mM KCl, 0.1% β‐mercaptoethanol, 1 mM Na3VO4, 5 mM NaF, 0.1 mM Pefabloc and 0.6% IGEPAL CA‐360). The mix was allowed to swell on ice for 30 min. After centrifugation, the supernatant was discarded and the pellet was then resuspended in a cold buffer B (20 mM HEPES, 2 mM EDTA, 420 mM NaCl, 0.1% β‐mercaptoethanol, 1 mM Na3VO4, 5 mM NaF, 0.1 mM Pefabloc) and incubated on ice for 30 min. Cellular debris was discarded by centrifugation and the supernatant fraction containing the nuclear fraction was stored. Protein concentration was determined by using the Bradford method (Panreac Applichem, Spain).
Equal amounts of protein were loaded onto polyacrylamide gels, transferred to PVDF membranes (Amersham‐GE Healthcare, UK) and probed with the following primary antibodies: anti‐glycerol kinase, anti‐NUR77, anti‐acetyl‐CoA carboxylase and anti‐phospho‐acetyl‐CoA carboxylase, anti‐HSP90, anti‐β‐actin and anti‐TBP (Table 2). After incubation with appropriate secondary antibodies conjugated with horseradish peroxidase, the immunoreactive bands were visualized using the enhanced chemiluminescence (ECL) system (GE‐Healthcare, England) according to the manufacturer's instructions and quantified by densitometry. HSP90 was used as a loading control for glycerol kinase and ACC. β‐actin was used as a loading control for NUR77. TBP was used as a loading control for glycerol kinase in the nuclear fraction.
TABLE 2.
Antibodies
| Target | Concentration | Reference |
|---|---|---|
| Glycerol kinase | 1:2000 | A6377, ABclonal |
| NUR77 | 1:1000 | A13316, ABclonal |
| Acetyl CoA carboxylase | 1:500 | 05‐1098, Merck |
| Phospho‐acetyl CoA carboxylase (Ser79) | 1:500 | 07‐303, Merck |
| HSP90 | 1:500 | 05‐594, Merck |
| β‐actin | 1:5000 | A5316, Merck |
| TBP | 1:1000 | T1827, Merck |
| Rabbit immunoglobulin HRP‐linked | 1:10 000 | A0545, Merck |
| Mouse immunoglobulin HRP‐linked | 1:10 000 | A9044, Merck |
2.6. Statistical analysis
Results are expressed as mean ± SEM. A Grubbs' test was run to detect outliers. Statistical comparisons between two groups were made using the unpaired Student's t‐test; when variances were not equal the analysis was made by the Mann–Whitney test. To compare body weight evolution and the increment of body weight, a two‐way ANOVA was performed using GraphPad Prism v8 (San Diego, CA, USA).
3. RESULTS
3.1. Ptn −/− mice have a lower weight gain during pregnancy
The evolution of the weight of pregnant mice from both genotypes was monitored daily. As shown in Figure 1A,B, Ptn −/− mice were smaller before pregnancy, had lower body weight during pregnancy (two‐way ANOVA F(9,250) for time and F(1,250) for genotype; p < .001 for all days) and, at the time of sacrifice they had lower weight gain than Ptn+/+ mice.
FIGURE 1.

Ptn −/− mice show decreased body weight and less weight gain during pregnancy. (A) Body weight evolution, (B) increase of the body weight during pregnancy, (C) conceptus weight (fetus‐placenta structure), and (D) retroperitoneal adipose tissue in Ptn+/+ (grey lines and bars) and Ptn−/− (blue lines and bars) pregnant mice. Data are expressed as mean ± SEM (n = 15–19 animals/group) *p < .05, **p < .01, ***p < .001 for differences between Ptn−/− and Ptn+/+ mice
Maternal body weight free of conceptus was also estimated. As shown in Figure 1C, the decrease in body weight of Ptn −/− pregnant mice, is due to a significant decrease both in the weight of maternal structures and the weight of conceptus (fetus‐placental structures). It is also worthy to highlight that Ptn −/− pregnant mice present a smaller retroperitoneal adipose tissue depot in comparison with the Ptn+/+ mice (Figure 1D).
3.2. Ptn deletion alters the plasma biochemical profile and exacerbates glucose intolerance during pregnancy
We next analyzed whether Ptn deletion could affect maternal biochemical parameters. As shown in Figure 2A, fasted glycemia was significantly higher in pregnant Ptn −/− mice than in controls and, although there were no differences in cholesterol or glycerol values, the circulating levels of NEFA and triacylglycerides were significantly lower in Ptn −/− pregnant mice than in Ptn+/+ (Figure 2B–E). The analysis by agarose electrophoresis of the different fractions of lipoproteins in plasma showed a significant decrease in both VLDL/IDL and LDL fractions in the Ptn −/− compared to the Ptn+/+ mice. No significant differences were observed in the HDL fraction (Figure 2F–H).
FIGURE 2.

Altered fasting plasma biochemical parameters, hormones, plasma lipoproteins, and glucose tolerance in vivo in 18 days pregnant Ptn −/− mice. (A) Glucose; (B) cholesterol; (C) glycerol, (D) non‐esterified fatty acids (NEFA), (E) triacylglycerides, (F) VLDL/IDL, (G) LDL, (H) HDL, (I) insulin, (J) glucagon, (K) glucose‐induced insulinotropic peptide (GIP), (L) glucose tolerance curve (GTT), (M) Area under the curve (AUC) for glucose during the GTT, and (N) HOMA‐IR. Data are expressed as mean ± SEM (n = 4–9 animals/group) *p < .05, **p < .01, and ***p < .001 for differences between Ptn−/− and Ptn+/+ mice
We also found that insulinemia was lower (Figure 2I), and circulating glucagon was significantly higher (Figure 2J) in pregnant Ptn −/− animals when compared to wild‐type mice, whereas no differences were found in the glucose‐induced insulinotropic peptide (GIP) levels (Figure 2K).
A glucose tolerance test (GTT) was performed on day 18 of gestation and after 6‐h fasting. As shown in the curves, glucose tolerance was significantly decreased (Figure 2L) and the area under the curve (AUC) was significantly increased in pregnant Ptn −/− mice compared to wild type animals (Figure 2M).
Given the lower glucose tolerance of the pregnant Ptn −/− mice, we next analyzed the HOMA‐IR index, an insulin resistance index. As shown in Figure 2N, no significant differences were observed between the two experimental groups.
3.3. Ptn −/− pregnant mice have a decreased hepatic lipid content
At day 18 of gestation, both the weight of the liver and the hepatic accumulation of lipids were lower in pregnant Ptn −/− than in wild type mice (Figure 3A,B). Moreover, when we determined the content of the different fractions respecting the total liver weight, a significant decrease was observed in all the lipid fractions in Ptn −/− mice compared to control animals (Figure 3C–F).
FIGURE 3.
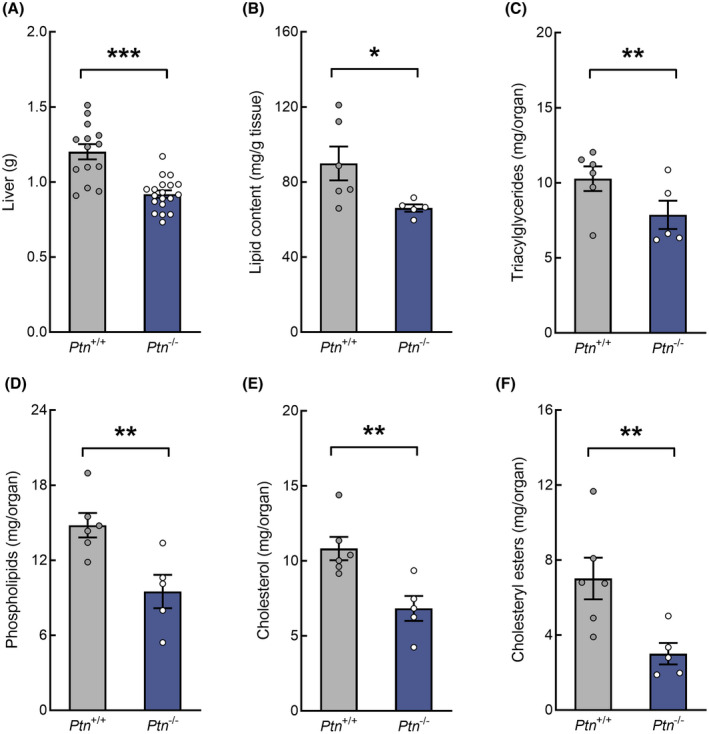
Effect of pleiotrophin deletion on liver weight, total lipid content, and hepatic lipid fractions at day 18 of pregnancy. (A) liver weight, (B) total lipid content, (C) triacylglycerides, (D) phospholipids, (E) cholesterol, and (F) cholesteryl esters. Data are expressed as mean ± SEM (n = 5–6 animals/group) *p < .05, **p < .01, and ***p < .001 for differences between Ptn −/− and Ptn +/+ mice
3.4. Ptn deletion impairs glucose synthesis and catabolism during late pregnancy
Glut2 expression in the liver was lower in pregnant Ptn −/− when compared to the control animals (Figure 4A). Moreover, pregnant Ptn −/− mice showed lower expression of glucokinase, phosphofructokinase I and pyruvate kinase (Figure 4B–D), suggesting that both the uptake of glucose and its transformation into pyruvate are reduced.
FIGURE 4.

Deletion of pleiotrophin impairs hepatic glucose metabolism in 18 days pregnant mice. (A) Glut2 mRNA, (B) Glucokinase (Gck) mRNA, (C) Phosphofructokinase I (Pfk1) mRNA, (D) Pyruvate kinase (Pk) mRNA, (E) Lactate dehydrogenase (Ldh) mRNA, (F) Pyruvate dehydrogenase α1 (Pdha1) mRNA, (G) Pyruvate carboxylase (Pc) mRNA, (H) Phosphoenolpyruvate carboxykinase (Pepck) mRNA, (I) Fructose 1,6 bisphosphatase (Fbp1) mRNA, (J) Glucose 6‐phosphatase (G6pc) mRNA, and (K) Glucose 6‐phosphate dehydrogenase (G6pd) mRNA. Data are expressed as mean ± SEM (n = 9 animals/group) *p < .05, **p < .01, and ***p < .001 for differences between Ptn −/− and Ptn +/+ mice
We also found that the mRNA expression of the enzymes responsible for the transformation of pyruvate into lactate (lactate dehydrogenase), acetyl‐CoA (pyruvate dehydrogenase) and oxaloacetate (pyruvate carboxylase) were decreased in the pregnant Ptn −/− mice when compared with the wild‐type pregnant mice (Figure 4E–G).
In addition, the analysis of the gluconeogenic enzymes phosphoenolpyruvate carboxykinase; fructose 1,6 bisphosphatase and glucose 6‐phosphatase revealed a two‐fold decrease in the mRNA expression (Figure 4H–J) in Ptn −/− pregnant mice when compared to the Ptn+/+ animals.
On the other hand, the expression of glucose 6‐phosphate dehydrogenase, the limiting enzyme of the pentose phosphate pathway, was increased in the Ptn −/− pregnant mice when compared with the control animals (Figure 4K).
3.5. Hepatic oxidation of fatty acids is decreased in Ptn −/− pregnant mice
We next analyzed the role of Ptn deletion in the oxidation of fatty acids. As shown in Figure 5A–D, the expression of carnitine palmitoyl transferase 1α, long chain acyl‐CoA dehydrogenase, very long chain acyl‐CoA dehydrogenase and 3‐hydroxyacyl‐CoA dehydrogenase are significantly decreased in Ptn −/− mice when compared to the wild‐type animals. Moreover, the expression of the peroxisomal acyl‐CoA oxidase 1 gene was also reduced in the Ptn −/− mice (Figure 5E).
FIGURE 5.

Impaired expression of enzymes involved in fatty acid oxidation in Ptn −/− 18 days pregnant mice. (A) Carnitine palmitoyl transferase 1α (Cpt1α) mRNA, (B) Acyl‐CoA dehydrogenase long chain (Acadl) mRNA, (C) Acyl‐CoA dehydrogenase very long chain (Acavdl) mRNA, (D) 3‐Hydroxyacyl‐Coa dehydrogenase α (Hadha) mRNA, and (E) Acyl‐CoA oxidase 1 (Acox1) mRNA. Data are expressed as mean ± SEM (n = 9 animals/group) *p < .05, **p < .01, and ***p < .001 for differences between Ptn −/− and Ptn +/+ mice
3.6. Hepatic fatty acids, triacylglycerides and cholesterol synthesis are decreased in Ptn −/− pregnant mice
In view of the changes described above, we determined the expression of the enzymes involved in hepatic lipid synthesis. First, Ptn −/− pregnant animals have a reduced mRNA expression of the enzymes involved in fatty acid synthesis (ATP citrate lyase, acetyl‐CoA carboxylase and fatty acid synthase) (Figure 6A–C) and desaturation (stearoyl‐CoA desaturase‐1) (Figure 6D). To confirm these results, the protein amount and the phosphorylation of acetyl‐CoA carboxylase, the limiting enzyme of lipogenesis, were determined by western blot. We found that not only the mRNA but also the acetyl‐CoA carboxylase protein levels were decreased in the liver of Ptn −/− when compared to Ptn +/+ pregnant mice (Figure 6B,M,O). Furthermore, the ratio phospho‐acetyl‐CoA carboxylase/acetyl‐CoA carboxylase (the inactive state of the enzyme) was significantly higher in these animals (Figure 6N,O), confirming that deletion of Ptn is associated with a significant increase in the inactive form of acetyl‐CoA carboxylase.
FIGURE 6.

At late pregnancy deletion of pleiotrophin is associated with changes in key enzymes involved in fatty acids, triacylglycerides and cholesterol synthesis. (A) ATP citrate lyase (Acly) mRNA, (B) Acetyl‐CoA carboxylase (Acc) mRNA, (C) Fatty acid synthase (Fas) mRNA, (D) Stearoyl‐CoA desaturase‐1 (Scd1) mRNA, (E) Lipin 2 (Lpin2) mRNA, (F) Glycerol phosphate acyltransferase (Gpat) mRNA, (G) Diacyglycerol acyltransferase‐1 (Dgat1) mRNA, (H) Diacyglycerol acyltransferase‐2 (Dgat2) mRNA, (I) Acetyl‐CoA acetyltransferase‐2 (Acat2) mRNA, (J) Hydroxymethylglutaryl‐CoA synthase (Hmgs) mRNA, (K) Hydroxymethylglutaryl‐CoA reductase (Hmgr) mRNA, (L) Sterol O‐acyltransferase (Soat) mRNA, (N) Acetyl‐CoA carboxylase total protein/HSP90, (M) ratio of phospho‐acetyl‐CoA carboxylase/acetyl‐CoA carboxylase protein, and (O) representative blots of phospho‐acetyl‐CoA carboxylase, acetyl‐CoA carboxylase, and HSP90. Data are expressed as mean ± SEM (n = 8–9 animals/group) *p < .05, **p < .01, and ***p < .001 for differences between Ptn −/− and Ptn +/+ mice
Moreover, as shown in Figure 6E–H, the hepatic mRNA expression of enzymes involved in triacylglycerol synthesis, lipin‐2, glycerol‐phosphate acyltransferase, diacylglycerol acyltransferase‐1, and diacylglycerol acyltransferase‐2 were also reduced in Ptn −/− as compared to Ptn +/+ pregnant mice. Finally, the mRNA of the key enzymes of cholesterol synthesis (acetyl‐CoA acetyltransferase‐2, hydroxymethylglutaryl‐CoA synthase, hydroxymethylglutaryl‐CoA reductase) (Figure 6I–K) and esterification (sterol O‐acyltransferase) (Figure 6L) were also diminished in Ptn −/− pregnant mice when compared to Ptn +/+ pregnant animals.
3.7. Membrane transporters and lipoprotein related enzymes are altered in the liver of Ptn −/− pregnant mice
We did not observe any changes in the expression of lipoprotein lipase mRNA between the two groups (Figure 7A). However, the expression of the hepatic triacylglyceride lipase was diminished in Ptn −/− mice (Figure 7B). The expression of the membrane transporters responsible for the hepatic uptake of glycerol and free fatty acids (Aqp9, Fatp4, and Fatp5) were significantly decreased in Ptn −/− when compared to Ptn +/+ animals (Figure 7C–E).
FIGURE 7.

Pleiotrophin deletion alters transporters and enzymes of lipoprotein metabolism in mice on day 18 of pregnancy. (A) Lipoprotein lipase (Lpl) mRNA, (B) Hepatic lipase (Lipc) mRNA, (C) Aquaporin 9 (Aqp9) mRNA, (D) Fatty acid transporter protein 4 (Fatp4) mRNA, (E) Fatty acid transporter protein 5 (Fatp5) mRNA, (F) Perilipin 2 (Plin2) mRNA, (G) Apo C‐II (ApoC2) mRNA, and (H) Apo B‐100 (ApoB100) mRNA. Data are expressed as mean ± SEM (n = 9 animals/group) *p < .05, **p < .01, and ***p < .001 for differences between Ptn −/− and Ptn +/+ mice
The expression of the most abundant lipid droplet protein in the liver, perilipin‐2 (Figure 7F), the hepatic levels of ApoC2 mRNA, a key component of VLDL and an activator of lipoprotein lipase (Figure 7G), and the expression of ApoB100 mRNA, the apolipoprotein necessary for the assembly and secretion of VLDL (Figure 7H) were also decreased in the pregnant mice in which Ptn expression has been knocked‐out.
Altogether, these results point out a clear reduction in fatty acid and glycerol uptake in the liver and may account for the reduction observed in the VLDL/IDL fraction (Figure 2F).
3.8. Ptn −/− pregnant mice show an impaired expression of transcription factors
Pregnant Ptn −/− mice showed downregulation of mRNA expression of Ppar‐α and Ppar‐γ1 (Figure 8A,B), whereas the expression of Ppar‐β/δ and the cofactor Pgc1α presented no variation (Figure 8B–D). In addition, the expression of Rxrα that is required for Ppar‐α transcriptional activity on β‐oxidation genes, and other important transcription factors involved in the regulation of liver metabolism, such as Srebp1, Fgf21, and Cebpα, were also diminished in Ptn −/− when compared to control pregnant mice (Figure 8E–H).
FIGURE 8.

Deletion of pleiotrophin is associated with differential expression of genes involved in the regulation of glucose and lipid metabolism in the liver of 18 days pregnant mice. (A) Ppar‐α mRNA, (B) Ppar‐γ1 mRNA, (C) Ppar‐βδ mRNA; (D) Pgc1‐α mRNA, (E) Rxr‐α mRNA, (F) Srebp1 mRNA, (G) Fgf21 mRNA, (H) Cebp‐α mRNA, (I) Nur77 mRNA, (J) NUR77 protein/β‐actin, (K) Glycerol kinase (Gyk) mRNA, (L) Glycerol kinase protein/HSP90, (M) Correlation analysis of glycerol kinase (Gyk) mRNA and Nur77 mRNA, (N) Glycerol kinase nuclear protein/TBP protein. Data are expressed as mean ± SEM (n = 3–9 animals/group) *p < .05, **p < .01, and ***p < .001 for differences between Ptn−/− and Ptn+/+ mice
NUR77 (mice isoform for NR4A1) is another transcription factor that has been implicated in the regulation of liver metabolism. As shown, its expression was significantly increased in the knock‐out mice when compared to the control pregnant animals (Figure 8I), although its protein level was found to be significantly reduced in Ptn −/− mice as compared to Ptn +/+ animals (Figure 8J). As glycerol kinase has been identified as a novel corepressor of NUR77 in the liver, we analyzed both the mRNA and protein of this enzyme and performed a correlation analysis of the levels of expression of both Nur77 and Gyk mRNA. Noteworthy, the mRNA and protein levels of glycerol kinase were significantly increased in Ptn −/− pregnant mice when compared to the wild‐type pregnant animals (Figure 8K,L). As shown in Figure 8M, Gyk mRNA has a significant positive correlation with the mRNA of Nur77 (r = .7836 and r = .6776 for Ptn +/+ and Ptn −/−, respectively; r = .7443 for all animals; p < .05 for all Pearson coefficients). Finally, we found that the abundance of glycerol kinase in the nuclei was significantly higher in Ptn −/− than in the Ptn +/+ pregnant mice (Figure 8N), suggesting that translocation of the enzyme to the nuclei is favored in the absence of PTN.
4. DISCUSSION
In this study, we provide novel insights into the role of pleiotrophin as a key player in the regulation of maternal metabolism during pregnancy. Pleiotrophin has been shown to be involved in the development, especially at early stages of differentiation, it is expressed in the placenta and its circulating levels are known to increase as pregnancy progresses. 22 Although pleiotrophin has shown to have a role in the regulation of glucose homeostasis and energy metabolism in non‐pregnant animals, 23 little is known about the functions of this protein in maternal metabolism during pregnancy.
Maternal liver plays a key role in the regulation of whole‐body metabolism during early pregnancy, 24 including the accumulation of fat depots. Additionally, during late pregnancy, the augmented lipolytic activity of adipose tissue favors the release of free fatty acids and glycerol into the circulation and these substrates are easily uptaken and esterified in the liver. Liver triacylglycerides are rapidly released into the circulation leading to the increase in the VLDL levels and the development of maternal hyperlipemia. 25
In this study, we show that Ptn deletion compromise maternal increment of body weight during pregnancy and this reduced body weight cannot be fully explained just by a decrease in the conceptus weight, as the maternal body weight free of conceptus in Ptn −/− mice is also lower than in wild type animals. Moreover, at late pregnancy, Ptn deletion was associated with a decrease in liver weight and hepatic lipid content. First, this data can be partially explained by the reduced expression of the enzymes involved in cholesterol synthesis and esterification. Second, Ptn deletion also reduced hepatic uptake of triacylglycerides from lipoproteins, as evidenced by the decreased expression of lipoprotein lipase and hepatic lipase, and by the lower expression of the main transporters involved in glycerol and fatty acids hepatic uptake (Aqp9, Fatp4, and Fatp5). Thirdly, deletion of Ptn is also associated with both a reduced expression of the key enzymes of lipogenesis, triacylglycerides synthesis and of perilipin 2, the main structural protein of lipid droplets that is also involved in the regulation of their dynamic and triacylglycerides metabolism. As a result of these impairments, Ptn deficiency is associated with an altered lipid metabolism that averts the normal lipid accumulation that takes place in the liver during pregnancy. Consequently, although it has been established that at late pregnancy circulating fatty acid and triacylglycerides levels are increased more than 250%, 26 , 27 in Ptn −/− pregnant mice the hepatic secretion of triacylglycerides as very‐low‐density lipoproteins (VLDL), and the circulating low‐density lipoproteins (LDL) and VLDL/IDL fractions are significantly reduced. Accordingly, the mRNA of apolipoprotein B100, which provides the structural framework required for the developing VLDL, and the mRNA of apolipoprotein C‐II, a cofactor of lipoprotein lipase, 28 are also decreased. In addition to these effects in the liver, we cannot discard other adaptations in extrahepatic tissues that may also contribute to the decrease in circulating lipids observed in the Ptn −/− pregnant mice. In this line of evidence, we have previously reported in this knock‐out mouse model an augmented energy production from fatty acids instead of glucose in extrahepatic tissues or increased fatty acid oxidation by thermogenesis. 23 Altogether, these results point out that, at late pregnancy, the deletion of Ptn is associated with a clear reduction in fatty acid, triacylglycerides and cholesterol synthesis in the liver that may explain the diminished hepatic lipid accumulation (Figure 9) as well as the decrease in circulating NEFA and triacylglycerides levels observed in these animals.
FIGURE 9.
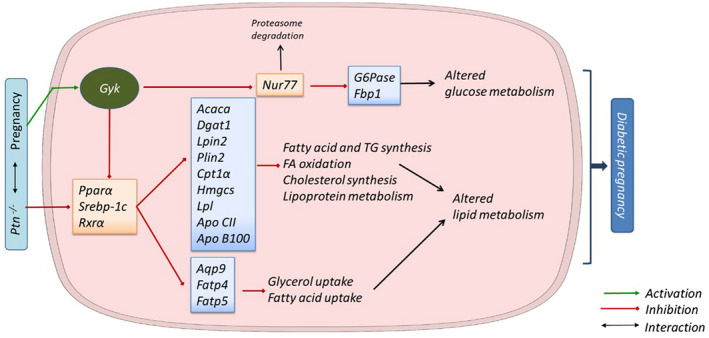
Summary of the molecular mechanism by which Ptn depletion alters liver metabolism during pregnancy, and the involvement of glycerol kinase as a regulatory factor. Pregnancy involves a series of metabolic changes that in turn can be affected by the deletion of the Ptn gene. In our study, Ptn −/− pregnant mice exhibit diminished glycerol and fatty acid uptake caused by a decrease in the expression of the transporters Aqp9, Fatp4, and Fatp5. Moreover, in the condition of lacking Ptn several transcription factors (PPARα, RXRα, SREBP‐1c) and key proteins involved in lipid metabolic pathways are downregulated, contributing to the reduction in lipid depots and circulating levels of triacylglycerides (TG) and NEFA. On the other hand, these factors are regulated by glycerol kinase, which links to carbohydrate metabolism. Gyk, which is upregulated in Ptn −/− pregnant mice, inhibits the transcriptional function of Nur77, a key regulator of glucose metabolism, including impaired gluconeogenesis. Both altered lipid and carbohydrate metabolism in the liver may favor the development of diabetes in pregnancy
We next explored whether Ptn deficiency was associated with the development of whole‐body glucose intolerance and/or altered glucose metabolism. Ptn −/− pregnant mice have hypoinsulinemia and are glucose intolerant, suggesting that Ptn deletion could favor a diabetogenic state in the pregnant dams. The decreased uptake and use of glucose by the liver in Ptn −/− pregnant mice might contribute to the maternal hyperglycemia, as the expression of the hepatic glucose transporter Glut2 and the glycolytic enzymes were reduced in late Ptn −/− pregnant mice (Figure 9). Furthermore, the increased insulin:glucagon ratio in Ptn −/− pregnant mice in the absence of a significant increase in GIP, may also contribute to the mobilization of stored nutrients, impairing fat accumulation and/or promoting the breakdown of adipose tissue depots. 29
It is known that most of the enzymes involved in glucose and lipid metabolism, including fatty acid transporters 30 are regulated by transcription factors as PPAR‐α, PPAR‐γ, RXR‐α, FGF21, and SREBP. 31 , 32 , 33 In fact, during the third trimester of pregnancy the decrease in the nuclear hormone receptor liposensors, including RXRs, PPARs or SREBP and the concomitant decrease in the expression of their target genes, has been proposed as one mechanism that leads to the alterations in lipid metabolism that characterize the third trimester of pregnancy. 34 We found that deletion of Ptn further decreases the expression of these transcription factors, and these may be involved in the altered lipid metabolism that we found in these animals.
Interestingly, the mRNA and protein of glycerol kinase were increased in the liver of these animals. 35 Recently, it has been shown that glycerol kinase apart from its kinase activity, transforming glycerol to glycerol 3‐phosphate in an ATP‐dependent reaction, has other functions, 36 , 37 including an essential role in lipid metabolism by affecting transcription factors activity, as SREBP‐1c or PPAR‐α. 35 Additionally, glycerol kinase is also a corepressor of NR4A1 (NUR77), a transcription factor that regulates hepatic glucose homeostasis. In fact, Gyk mRNA overexpression has been shown to suppress NR4A1 ability to regulate the expression of hepatic gluconeogenic genes, such as glucose 6 phosphatase and fructose 1,6‐bisphosphatase, and to modulate hepatic glucose production in the unfed state and diabetes in vitro and in vivo. 36 , 38 In this scenario, the increased amount of glycerol kinase in late pregnant Ptn −/− mice may suppress the transcriptional activity of NUR77, by attenuating its binding to gluconeogenic gene promoters, negatively regulating their transcription as has been found in other studies. 36 Supporting this role of glycerol kinase, we found that deletion of Ptn is associated with an increased translocation of glycerol kinase into the nuclei of liver cells. Additionally, the proteasome degradation of NUR77 may be enhanced (Figure 9) as undetectable protein levels were found in the liver of Ptn −/− pregnant mice. In line with this hypothesis, it has been shown that NUR77 degradation is controlled by SUMOylation and later ubiquitination, and that for the SUMOylation of NUR77 the protein must be previously phosphorylated. 39 , 40
In view of these results, it is tempting to speculate that deletion of Ptn may be associated in the liver with altered transcriptional activity modulated by glycerol kinase, impairing the pregnancy‐related adaptations in both glucose and lipid metabolism, including glucose intolerance and a decreased capacity to store lipids (Figure 9).
The mouse is an important mammalian model to approach the study of human metabolism in pregnancy, particularly at a molecular level. Although the present study has some limitations, as we cannot discard the contributions of other organs, such as adipose tissue, to the observed changes in lipid metabolism in liver, we suggest that this model will be valuable to elucidate the mechanisms underlying adaptations of liver metabolism during obese pregnancy, which is frequently associated to fatty liver. In fact, fatty liver is rather common in human pregnancy, because of increasing prevalence of diabetes and/or obesity, and it is associated with adverse maternal and perinatal outcomes. 41 Thus, future studies of pregnant Ptn −/− mice fed an obesogenic diet, will provide additional information on the maternal factors that may protect pregnant mothers to develop fatty liver disease.
5. CONCLUSION
Our study is the first one demonstrating the key role of pleiotrophin in the maintenance of hepatic homeostasis during pregnancy. Deletion of Ptn renders mice with altered metabolism of both glucose and lipids, which is associated with a diabetogenic state in the pregnant mother. The finding that Ptn deficiency alters the expression profile of a set of genes regulating lipid uptake and glucose and lipid utilization, supports the notion that defective PPAR‐α and NUR77 activation accounts for the phenotype of our mouse model, by inducing impairments in lipid and glucose homeostasis (Figure 8), and further highlighted the moonlighting role of glycerol kinase in pregnancy.
DISCLOSURES
The authors declared that no conflicts of interest exist with this manuscript.
AUTHOR CONTRIBUTIONS
The experimental work was performed in the research laboratory of MPR‐A in the Department of Chemistry and Biochemistry, Universidad San Pablo‐CEU. All authors have contributed to the conception or design of the work, acquisition or analysis or interpretation of data for the work, drafting the work or revising it critically for important intellectual content, approved the final version to be published and agreed to be accountable for all aspects of the work.
ACKNOWLEDGEMENTS
We thank Monica Diez Hochleitner‐Ruiz for the analysis of some circulating parameters, and our colleagues in the Animal Facility of Universidad San Pablo‐CEU. J Sevillano, MS Sánchez‐Alonso and B Zapatería were recipients of CEU‐Santander mobility grants. This work was supported by the Spanish Ministry of Science and Innovation (RTI2018‐095615‐B‐I00) and the Community of Madrid (S2017/BMD‐3684) to MP Ramos‐Alvarez.
Zapatería B, Sevillano J, Sánchez‐Alonso MG, et al. Deletion of pleiotrophin impairs glucose tolerance and liver metabolism in pregnant mice: Moonlighting role of glycerol kinase. FASEB J. 2021;35:e21911. 10.1096/fj.202101181R
REFERENCES
- 1. Catalano PM. Obesity, insulin resistance, and pregnancy outcome. Reproduction. 2010;140:365‐371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Herrera E. Metabolic adaptations in pregnancy and their implications for the availability of substrates to the fetus. Eur J Clin Nutr. 2000;54(Suppl 1):S47‐S51. [DOI] [PubMed] [Google Scholar]
- 3. Ramos MP, Crespo‐Solans MD, del Campo S, Cacho J, Herrera E. Fat accumulation in the rat during early pregnancy is modulated by enhanced insulin responsiveness. Am J Physiol Endocrinol Metab. 2003;285:E318‐E328. [DOI] [PubMed] [Google Scholar]
- 4. Herrera E, del Campo S, Marciniak J, Sevillano J, Ramos MP. Enhanced utilization of glycerol for glyceride synthesis in isolated adipocytes from early pregnant rats. J Physiol Biochem. 2010;66:245‐253. [DOI] [PubMed] [Google Scholar]
- 5. Villar J, Cogswell M, Kestler E, Castillo P, Menendez R, Repke JT. Effect of fat and fat‐free mass deposition during pregnancy on birth weight. Am J Obstet Gynecol. 1992;167:1344‐1352. [DOI] [PubMed] [Google Scholar]
- 6. López‐Luna P, Maier I, Herrera E. Carcass and tissue fat content in the pregnant rat. Biol Neonate. 1991;60:29‐38. [DOI] [PubMed] [Google Scholar]
- 7. López‐Luna P, Muñoz T, Herrera E. Body fat in pregnant rats at mid‐ and late‐gestation. Life Sci. 1986;39:1389‐1393. [DOI] [PubMed] [Google Scholar]
- 8. Herrera E. Lipid metabolism in pregnancy and its consequences in the fetus and newborn. Endocrine. 2002;19:43‐55. [DOI] [PubMed] [Google Scholar]
- 9. Carrara MA, Batista MR, Saruhashi TR, Felisberto AM Jr, Guilhermetti M, Bazotte RB. Coexistence of insulin resistance and increased glucose tolerance in pregnant rats: a physiological mechanism for glucose maintenance. Life Sci. 2012;90:831‐837. [DOI] [PubMed] [Google Scholar]
- 10. Herrera E, Muñoz C, López‐Luna P, Ramos P. Carbohydrate‐lipid interactions during gestation and their control by insulin. Braz J Med Biol Res. 1994;27:2499‐2519. [PubMed] [Google Scholar]
- 11. Krellman JW, Ruiz HH, Marciano VA, Mondrow B, Croll SD. Behavioral and neuroanatomical abnormalities in pleiotrophin knockout mice. PLoS ONE. 2014;9:e100597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gu D, Yu B, Zhao C, et al. The effect of pleiotrophin signaling on adipogenesis. FEBS Lett. 2007;581:382‐388. [DOI] [PubMed] [Google Scholar]
- 13. Xu C, Zhu S, Wu M, Han W, Yu Y. Functional receptors and intracellular signal pathways of midkine (MK) and pleiotrophin (PTN). Biol Pharm Bull. 2014;37:511‐520. [DOI] [PubMed] [Google Scholar]
- 14. Asahina K, Sato H, Yamasaki C, et al. Pleiotrophin/heparin‐binding growth‐associated molecule as a mitogen of rat hepatocytes and its role in regeneration and development of liver. Am J Pathol. 2002;160:2191‐2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sato H, Funahashi M, Kristensen DB, Tateno C, Yoshizato K. Pleiotrophin as a Swiss 3T3 cell‐derived potent mitogen for adult rat hepatocytes. Exp Cell Res. 1999;246:152‐164. [DOI] [PubMed] [Google Scholar]
- 16. Michelotti GA, Tucker A, Swiderska‐Syn M, et al. Pleiotrophin regulates the ductular reaction by controlling the migration of cells in liver progenitor niches. Gut. 2016;65:683‐692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ochiai K, Muramatsu H, Yamamoto S, Ando H, Muramatsu T. The role of midkine and pleiotrophin in liver regeneration. Liver Int. 2004;24:484‐491. [DOI] [PubMed] [Google Scholar]
- 18. Amet LE, Lauri SE, Hienola A, et al. Enhanced hippocampal long‐term potentiation in mice lacking heparin‐binding growth‐associated molecule. Mol Cell Neurosci. 2001;17:1014‐1024. [DOI] [PubMed] [Google Scholar]
- 19. Herradon G, Ezquerra L, Nguyen T, et al. Pleiotrophin is an important regulator of the renin‐angiotensin system in mouse aorta. Biochem Biophys Res Comm. 2004;324:1041‐1047. [DOI] [PubMed] [Google Scholar]
- 20. Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497‐509. [PubMed] [Google Scholar]
- 21. Ruiz JI, Ochoa B. Quantification in the subnanomolar range of phospholipids and neutral lipids by monodimensional thin‐layer chromatography and image analysis. J Lipid Res. 1997;38:1482‐1489. [PubMed] [Google Scholar]
- 22. Ball M, Carmody M, Wynne F, et al. Expression of pleiotrophin and its receptors in human placenta suggests roles in trophoblast life cycle and angiogenesis. Placenta. 2009;30:649‐653. [DOI] [PubMed] [Google Scholar]
- 23. Sevillano J, Sánchez‐Alonso MG, Zapatería B, et al. Pleiotrophin deletion alters glucose homeostasis, energy metabolism and brown fat thermogenic function in mice. Diabetologia. 2019;62:123‐135. [DOI] [PubMed] [Google Scholar]
- 24. Torres N, Bautista CJ, Tovar AR, et al. Protein restriction during pregnancy affects maternal liver lipid metabolism and fetal brain lipid composition in the rat. Am J Physiol Endocrinol Metab. 2010;298:E270‐E277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Herrera E, Lasunción MA, Gomez‐Coronado D, Aranda P, López‐Luna P, Maier I. Role of lipoprotein lipase activity on lipoprotein metabolism and the fate of circulating triglycerides in pregnancy. Am J Obstet Gynecol. 1988;158:1575‐1583. [DOI] [PubMed] [Google Scholar]
- 26. Darmady JM, Postle AD. Lipid metabolism in pregnancy. Br J Obstet Gynaecol. 1982;89:211‐215. [DOI] [PubMed] [Google Scholar]
- 27. Haggarty P. Fatty acid supply to the human fetus. Annu Rev Nutr. 2010;30:237‐255. [DOI] [PubMed] [Google Scholar]
- 28. Wolska A, Dunbar RL, Freeman LA, et al. Apolipoprotein C‐II: new findings related to genetics, biochemistry, and role in triglyceride metabolism. Atherosclerosis. 2017;267:49‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kalra S, Gupta Y. The insulin: glucagon ratio and the choice of glucose‐lowering drugs. Diabetes Ther. 2016;7:1‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ambühl P, Amemiya M, Preisig PA, Moe OW, Alpern RJ. Chronic hyperosmolality increases NHE3 activity in OKP cells. J Clin Investig. 1998;101:170‐177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Horton JD, Shah NA, Warrington JA, et al. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci U S A. 2003;100:12027‐12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rakhshandehroo M, Knoch B, Müller M, Kersten S. Peroxisome proliferator‐activated receptor alpha target genes. PPAR Res. 2010;2010:1‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang Y, Nakajima T, Gonzalez FJ, Tanaka N. PPARs as metabolic regulators in the liver: lessons from liver‐specific PPAR‐null mice. Int J Mol Sci. 2020;21(6):2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sweeney TR, Moser AH, Shigenaga JK, Grunfeld C, Feingold KR. Decreased nuclear hormone receptor expression in the livers of mice in late pregnancy. Am J Physiol Endocrinol Metab. 2006;290:E1313‐E1320. [DOI] [PubMed] [Google Scholar]
- 35. MacLennan NK, Rahib L, Shin C, et al. Targeted disruption of glycerol kinase gene in mice: expression analysis in liver shows alterations in network partners related to glycerol kinase activity. Hum Mol Genet. 2006;15:405‐415. [DOI] [PubMed] [Google Scholar]
- 36. Miao L, Yang Y, Liu Y, et al. Glycerol kinase interacts with nuclear receptor NR4A1 and regulates glucose metabolism in the liver. FASEB J. 2019;33:6736‐6747. [DOI] [PubMed] [Google Scholar]
- 37. Sriram G, Parr LS, Rahib L, Liao JC, Dipple KM. Moonlighting function of glycerol kinase causes systems‐level changes in rat hepatoma cells. Metab Eng. 2010;12:332‐340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pei L, Waki H, Vaitheesvaran B, Wilpitz DC, Kurland IJ, Tontonoz P. NR4A orphan nuclear receptors are transcriptional regulators of hepatic glucose metabolism. Nat Med. 2006;12:1048‐1055. [DOI] [PubMed] [Google Scholar]
- 39. Zárraga‐Granados G, Muciño‐Hernández G, Sánchez‐Carbente MR, et al. The nuclear receptor NR4A1 is regulated by SUMO modification to induce autophagic cell death. PLoS ONE. 2020;15:e0222072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang L, Xie F, Zhang J, Dijke PT, Zhou F. SUMO‐triggered ubiquitination of NR4A1 controls macrophage cell death. Cell Death Differ. 2017;24:1530‐1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sarkar M, Grab J, Dodge JL, et al. Non‐alcoholic fatty liver disease in pregnancy is associated with adverse maternal and perinatal outcomes. J Hepatol. 2020;73:516‐522. [DOI] [PMC free article] [PubMed] [Google Scholar]


