Abstract
Deletions and point mutations in the dystrophin gene cause either the severe progressive myopathy Duchenne muscular dystrophy (DMD) or the milder Becker muscular dystrophy, depending on whether the translational reading frame is lost or maintained. Because internal in-frame deletions in the protein produce only mild myopathic symptoms, it should be possible, by preventing the inclusion of specific mutated exon(s) in the mature dystrophin mRNA, to restore a partially corrected phenotype. Such control has been previously accomplished by the use of synthetic oligonucleotides; nevertheless, a significant drawback to this approach is caused by the fact that oligonucleotides would require periodic administrations. To circumvent this problem, we have produced several constructs able to express in vivo, in a stable fashion, large amounts of chimeric RNAs containing antisense sequences. In this paper we show that antisense molecules against exon 51 splice junctions are able to direct skipping of this exon in the human DMD deletion 48–50 and to rescue dystrophin synthesis. We also show that the highest skipping activity was found when antisense constructs against the 5′ and 3′ splice sites are coexpressed in the same cell.
Duchenne muscular dystrophy (DMD) is an X-linked recessive disorder that affects 1 in every 3,500 males. It is characterized by the absence of the cytoskeletal dystrophin (427-kDa protein) that in turn produces a severe and progressive muscle deterioration. Most of the DMD mutations consist in deletions and point mutations in the 2.5-Mb dystrophin gene that introduce stop codons and consequently premature translation termination. A milder myopathy is the Becker muscular dystrophy; in this case, deletions inside the gene produce in frame mRNAs and consequently shorter but semifunctional dystrophin proteins (1). A third of DMD cases are the results of a de novo mutation (2, 3), therefore the disease can never be eliminated through genetic screening and counseling. For this reason, many efforts are now being devoted to the development of a treatment for this disorder, and several strategies have been designed that might provide an insight into finding a cure. One of these strategies involves the transplantation of normal myoblasts into the muscle tissues that lack this protein (4, 5), whereas the other strategy tries to restore correct expression of the dystrophin through a gene therapy approach. In this direction, several groups have tried to deliver full-length or mini cDNA copies of dystrophin into cells with the mutated gene (6–8). Even if this approach is very promising, several problems still remain to be solved, such as size capacity and transducing activity of the vector and immune response to the “therapeutic” gene (9).
Another powerful approach of gene therapy is based on the fact that internal in-frame deletions in the protein produce only mild myopathic symptoms; therefore, it should be possible, by preventing the inclusion of specific mutated exon(s) in the mature dystrophin mRNA, to restore a partially corrected phenotype. In this respect, it is interesting to note that restoration of dystrophin levels as low as 30% are sufficient for substantial therapeutic benefit (10).
Exon-specific skipping has been accomplished by the extracellular delivery of synthetic 2′-O-methyl antisense oligonucleotides raised against specific splicing junctions (11–13) or exonic enhancers (10). The interaction of the antisense RNA with the corresponding target sequence should mask the utilization of specific splice sites or prevent the binding of enhancer factors during the splicing reaction and determine the skipping of the neighboring mutated exon(s) so as to produce an in-frame dystrophin mRNA, the ultimate effect being the production of a shorter but functional dystrophin protein. A significant drawback to the synthetic oligonucleotide approach is that it would require periodic administrations. To circumvent this problem, we have concentrated our efforts in the design and construction of vectors able to express in vivo, in a stable and continuous fashion, large amounts of chimeric RNAs containing antisense sequences. This strategy was tested on the human deletion of exons 48–50. In this case, a premature termination codon is produced in exon 51; if skipping of this exon is obtained the result would be an in-frame mRNA. We have used different small nuclear RNAs (snRNAs) and their corresponding genes to express in the nucleus chimeric molecules carrying antisense sequences complementary to exon 51 splice junctions, and we have tested their activity in DMD myoblast. The results obtained showed that the combination of antisense molecules against the 5′ and 3′ splice junctions induced efficient skipping of exon 51 and partial rescue of dystrophin synthesis.
Materials and Methods
Plasmid Constructs.
Clone U1–5′ was obtained by inverse PCR on plasmid pHU1-ID, containing the entire U1 RNA human gene, with the oligos: ANTI 24+2A, 5′-GTATGAGAAAAAATGAGATCTTGGGCCTCTGC-3′ and ANTI 24+2B 5′-CTTCTGCTTGATGGCAGGGGAGATACCATGATC-3′. The U1 expression cassette was constructed by inverse PCR, with oligonucleotides: U1cas A, 5′-CCTGGTACCATGAGATCTTGGGCCTCTGC-3′ and U1cas B, 5′-CCTCTCGAGCTGACTTTCTGGAGTTTC-3′. In this construct, the U1 snRNA coding region (from nt 3 to 161) was replaced with the KpnI, StuI, and XhoI restriction sites.
Clone U7–3′ was obtained by direct PCR on the U7SmOPT plasmid, kindly provided by D. Suter (12), with oligos: Amp, 5′-CCATAGTTGCCTGACTCCCCGTCGTG-3′ and U7F, 5′- ACAGAGGCCTTTCCGCAAGTCTGAGTAGGAGCT-AAAATATTTTGGGAATTTTTGGAGCAGG. The U7–3′ coding region was put under the control of the U1 promoter (construct U7–3′p1) by PCR amplification of U7–3′ DNA with oligos: U7down, 5′-CCGCTCGAGGGGTTTTCCGACCGAAG-3′ and U7up 5′-CGGGGTACCAAGTCTGAGTAGGAGCTAAA-3′. Construct U7-double is obtained by PCR amplification of U7–3′ with oligos U7down and U7d 5′- CGGGGTACCAATTTTTCTCATACCTTCTGCTTGATGTCTGAGTAGGAGCTAAAAT-3′ and cloned in the U1-expressing cassette after KpnI/XhoI digestion. After KpnI and XhoI digestion the amplified DNAs were cloned into the U1 expression cassette giving rise to U7–3′p1 and U7-double.
The human U2snRNA gene, U2h (kindly provided by A. Weiner), including the promoter and the 3′ downstream sequences, was used to generate by inverse PCR the U2mod construct with oligonucleotides U2mA, 5′-AAAGGCTGTCTTCGCATGCGCTCGCCTTCGCGCCC-3′ and U2mB, 5′-TGGCTATCTCTCGAGTATCAGTTTAATATCTGATAC-3′. The U2–BP construct was derived from U2mod by inverse PCR with oligonucleotides U2X, 5′-CTCGAGAGATAGCCAAAAGGCTGTCTTCGCATGCGCTCGCC-3′ and U2BP, 5′-AAAAAGAAGAAAAAGAAAAATTAGAAACTA-TCAGTTTAATATCTGATACG-3′.
Retroviral Constructs.
Single antisense constructs.
U7–3′ was amplified by direct PCR using oligonucleotides: U7cas up 5′-CTAGCTAGCCCACATCGCCTGCCACTAC-3′ and U7cas down 5′-CTAGCTAGCCCAGAGGAGGCAGAAAGG-3′ and U2BP with oligos: U2cas up 5′-CTAGCTAGCCAGGCCTTCGGCTTCCCTGACTGGG-3′ and U2cas down 5′-CTAGCTAGCGCGCGTCACAGGACTCGTGCAAGCC-3′. The resulting fragments were cloned in the forward orientation into the NheI site of the pBabe puro retroviral vector (14). The BamHI fragments containing the entire chimeric constructs U1–5′, U7–3′p1 and U7-double, were inserted in the pBabe puro NheI site modified with BglII adaptors.
Double antisense constructs.
To generate the 5′/3′ construct U1–5′ was amplified by direct PCR with oligos: U1cas up 5′- CTAGCTAGCGGTAAGGACCAGCTTCTTTG-3′ and U1cas down 5′-CTAGCTAGCGGTTAGCGTACAGTCTAC-3′. After NheI digestion the DNA fragment was cloned into the XbaI site of the U7-3′p1 plasmid. Digestion with HincII and SmaI produces a fragment containing both coding sequences. This molecule was inserted in the repaired NheI site of the pBabe puro vector. The 5′/BP construct was made by cloning the BamHI fragment containing the U1-5′coding region inside the U2-BP plasmid. This construct was amplified with oligos U2cas up and U1cas down and cloned in the NheI site of the pBabe vector.
Packaging Cell Line.
The Phoenix packaging cell line (http://www.stanford.edu/nolan) was cultured in DMEM supplemented with 10% FCS and 1% penicillin/streptomycin (GIBCO/BRL). Retroviral particles were collected from Phoenix cells transiently transfected with 4 μg of the various pBabe puro constructs by using CaPO4 and were used to transduce the Duchenne cell line.
Cell Culture.
The Duchenne derived biopsy (491A) was put in culture and maintained in RPMI medium 1640 supplemented with 15% FCS, 1% penicillin–streptomycin and 1% glutamine (GIBCO/BRL). Cells were propagated by standard trypsinization and seeded directly in a collagen-coated dish (rat tail collagen type 1, Becton Dickinson). Infection of the cells was carried out in the absence of any antibiotic. One day before infection, cells were plated at a density of ≈5 × 105 cells per 100-mm dish. Three rounds of infections were performed with the different viral supernatants. Forty-eight hours after infection, cells were divided and puromycin was added at the final concentration of 1 μg/ml (Sigma).
RNA Preparation and Analysis.
Total cellular RNA was prepared by using the Ultraspec RNA isolation system (Biotecx Laboratories, Houston). For Northern blot analysis, 8 μg of total RNA was loaded on 6% polyacrylamide/7 M urea gels, blotted, and hybridized with the following antisense specific probes: U1a 5′-ATCAAGCAGAAGGTATGAGAAAAA-3′; U2a 5′-GTTTCTAATTTTTCTTTTTCTTC-3′;U7a 5′-CCCAAAATATTTTAGCTCCTACTCAGAC-3′. Reverse transcription (RT)-PCR was carried out with 200 ng of total RNA for 40 cycles of amplification using Access RT-PCR system (Promega). Two microliter of the RT-PCR product was used as template in a 50-μl secondary nested PCR that was carried out for 30 cycles. Products were analyzed on 2.5% agarose gel, and specific bands were purified for sequence analysis using the QIAquick Gel extraction kit (Qiagen). DNA sequencing was carried out by M-Medical using the BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems) and analyzed on an ABI Prism 377 Sequencer.
Xenopus laevis Oocyte Microinjections.
AdML pre-mRNA is 253 nucleotides long and covers part of the major late transcript of adenovirus; it is obtained by in vitro transcription of plasmid pAdML-Di1 digested with ScaI (15). 32P-labeled AdML and U6 RNAs (respectively 1.5 × 106 and 0.4 × 106 cpm/μl) were coinjected in oocytes that had received the night before the U2-BP or U2h plasmids (4 ng per oocyte). Incubation was allowed to proceed for an additional hour; RNA was extracted and analyzed on a 6% polyacrylamide/7 M urea gel.
Western Blotting Analysis.
Cultured cells were washed twice with ice-cold PBS and then lysed for 40 min at 4°C in a buffer containing 500 mM Tris⋅HCl (pH 8), 150 mM NaCl, 100 mM NaF, 1 mM EGTA, 1.5 mM MgCl2, 1% Triton X-100, and 10% glycerol, and supplemented with various protease inhibitors (leupeptin, PMSF, apoprotein). After clearing by centrifugation at 15,300 × g for 10 min, protein content was quantified by Bio-Rad Protein Assay. 100 μg of protein was separated onto a 6% polyacrylamide gel containing 0.1% SDS and 150 mM Tris⋅HCl (pH 8.8), and transferred to polyscreen poly(vinylidene difluoride) ransfer membrane (Nen Life Science). After treatment with 5% nonfat dry milk in TBS-T (20 mM Tris·HCl, pH 7.8/136 mM NaCr/0.05% Tween 20), membrane was incubated with NCL-DYS1 mouse monoclonal antibody (Novocastra Laboratories) diluted 1:100 for 60 min, followed by three washes in TBS-T buffer and incubation with a horseradish peroxidase-linked secondary antibody (1:1,000; Amersham Pharmacia) for 45 min. Detection of proteins was carried out with super signal chemiluminescent substrate (Pierce).
Results
Construction of the Antisense Chimeric RNAs.
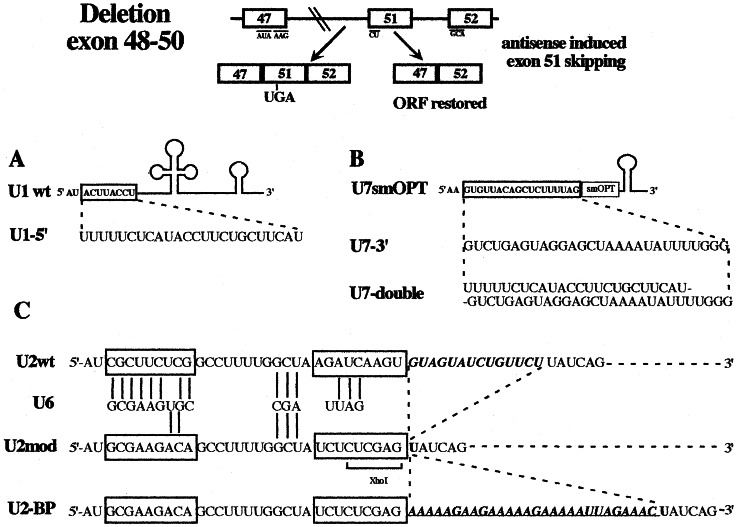
Among the series of cases made available to us, we selected one case, patient 491A, having a deletion encompassing exons 48, 49, and 50 (E.R. and G.G., unpublished data). In this case, skipping of exon 51 would produce an in frame mRNA coding for a protein 210 aa shorter than the wild-type dystrophin (see schematic representation of Fig. 1).
Figure 1.
Schematic representation of the dystrophin mutation used. Patient 491A has a deletion encompassing exon 48 through 50. Splicing of the pre-mRNA produces an out-of-frame mRNA with a stop codon in exon 51. The exclusion of exon 51 would instead produce an in-frame mRNA. (A) The U1–5′ construct was obtained by replacing the 5′ end (boxed nucleotides) of wild-type U1 snRNA with the antisense sequence shown underneath. (B) The U7smOPT (18, 19) was modified by replacing the region pairing with the histone pre-mRNA (boxed nucleotides) with a 28-nt-long sequence complementary to the region across intron 50/exon 51 (construct U7–3′). U7-double contains a longer substitution that combines the antisense element contained in U1–5′ with that of U7–3′. (C) The U2snRNA was initially converted into the U2mod construct that was in turn modified to give U2-BP. The sequence of the 5′ portion of the wild-type and U2 snRNA derivatives (nucleotides 1–52) is shown together with that of U6 snRNA complementary nucleotides. The boxed regions indicate the regions substituted, whereas nucleotides pairing to U6 are indicated by bars. The 14-nt-long region of U2, indicated in italics, containing the branch-site pairing region, has been deleted in U2 mod. In U2-BP this region was replaced with a 28-nt-long sequence complementary to the 3′ region, containing the branch site, of intron 50 (underlined sequence).
The U1 snRNA was engineered so as to substitute 8 nucleotides at its 5′ terminus with a 24-nt-long sequence complementary to the corresponding region across the splice junction of exon/intron 51 (construct U1–5′, see Fig. 1A). This choice was dictated by the fact that the 5′-terminal region of U1 snRNA is normally in a single-stranded conformation and is involved in the recognition of the 5′ splice junction of pre-mRNAs (16). The expression of the chimeric RNA is directed by a strong DNA polymerase II (polII)-dependent promoter (17).
A second series of constructs were raised in the U7 snRNA. We started from the U7 smOPT derived by Gorman et al. (18) by substituting the original suboptimal Sm-binding site of the wild-type U7 snRNA gene with a canonical sequence. This construct has higher stability if compared with the parental construct (19). The 5′ end region of U7 snRNA functions as a natural antisense sequence by hybridizing with the so-called spacer element of histone pre-mRNA during its 3′ processing (20–22). This region was then substituted with 28 nucleotides complementary to the 3′ splice junction of intron 50/exon 51 (construct U7–3′, see Fig. 1B). Because the polII-dependent promoter of the U7 gene was reported to have low transcriptional activity, we cloned U7–3′ under the U1 snRNA gene promoter, giving rise to construct U7–3′p1. A derivative of this last construct contains a second antisense sequence complementary to the 5′ splice junction of intron 51 (construct U7-double, see Fig. 1B). The second antisense sequence is the same as the one present in construct U1–5′ (Fig. 1A).
The last type of RNA vector used is the U2 snRNA. This RNA is normally involved in the splicing reaction by recognizing the intronic branch site sequence and directing the entry, in the spliceosome, of the catalytic U6 snRNA (23, 24). Both interactions are mediated by base pairing. In our design, we have first produced a U2 derivative (Fig. 1C, U2mod) with mutations in the region pairing to U6 snRNA. Nucleotide substitutions were made in such a way that the secondary structure of the U2 snRNA would not be affected. The lack of interaction with the U6 snRNA should ensure that the modified U2 snRNA would not interfere with the cellular splicing apparatus and would not recruit the spliceosome on the intron to which it is paired. U2 mod was subsequently modified so that the branch site pairing region was substituted with 28 nucleotides complementary to the branch site and 3′ region of intron 50, giving rise to construct U2-BP (Fig. 1C).
All of the described constructs were cloned separately or in couples in the 3′ LTR (U3 region) of the pBabe puro retroviral vector and viral particles were obtained by transfecting plasmid DNAs into the Phoenix packaging cell line (14).
Transduction of Antisense Constructs into DMD-Derived Cells and Analysis of Their Expression.
From a muscular biopsy of the 491A DMD patient, cells were put in culture and immortalized by retroviral-mediated integration of a wild-type SV40 large-T antigen according to Berghella et al. (25). Immortalized cells were infected with recombinant retroviral particles, and stable integrations were selected in puromycin-containing medium.
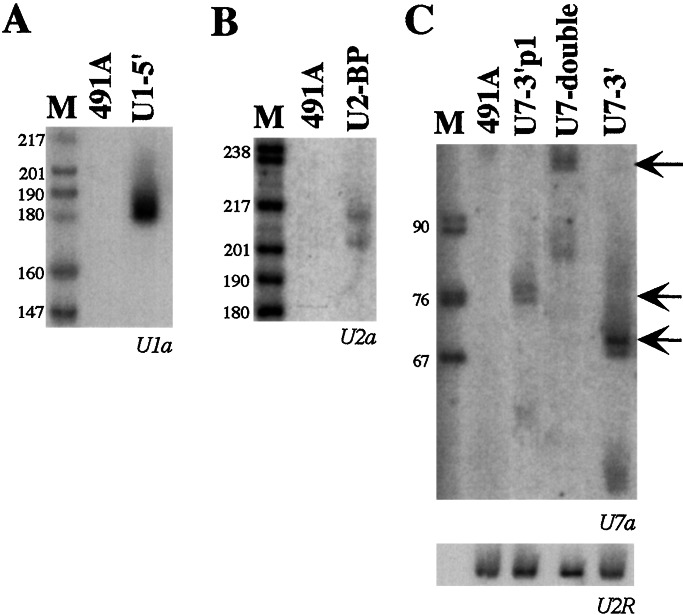
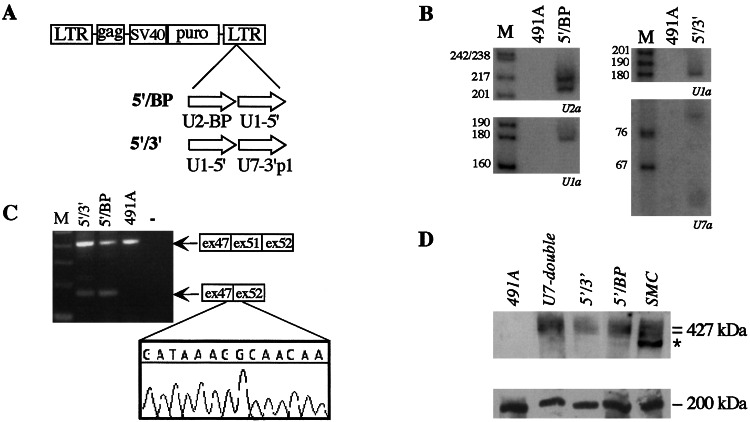
Expression of the antisense RNAs was checked by Northern analysis on the total cell population grown in the presence of puromycin. Fig. 2 shows that all of the constructs express the chimeric RNAs at fairly good levels.
Figure 2.
In vivo expression of the antisense constructs. Eight micrograms of total RNA, extracted from mixed cell populations transduced with the different chimeric pBabe puro vectors (indicated above the lanes) and from control untransduced cells (lanes 491A), were run on 6% polyacrylamide/7 M urea gels and electroblotted onto nylon membranes. Hybridizations were carried out with 32P-labeled oligonucleotides, specific for each antisense sequence, as indicated below each panel. To normalize the sample, the filter in C was also probed with the oligonucleotide U2R specific for endogenous U2snRNA. Arrows in C indicate the full-length products of the U7 derivatives. Lane M, pBR322 plasmid DNA, digested with MspI.
The U7 constructs (Fig. 2C) display the accumulation of full-length molecules together with a small amount of shorter products. Hybridizations with 5′- and 3′-specific probes showed that the shorter forms miss 20 nucleotides in the 3′ end of the molecule. It is not presently known whether the two types of transcripts have different stability; nevertheless, it appears that, under steady state conditions, mainly full-length products accumulate. Control hybridization on the endogenous U2 snRNA (Fig. 2C Lower) indicates that the highest accumulation was obtained when the chimeric U7 snRNAs were expressed under their own promoter.
Microinjection experiments into X. laevis oocytes, which easily allow to study the nucleus/cytoplasm localization, indicated that all of the constructs described above were expressed at high levels and specifically localized in the nuclear compartment (not shown). These results demonstrated that the modifications introduced in the different snRNAs did not affect either their overall stability or their subcellular localization.
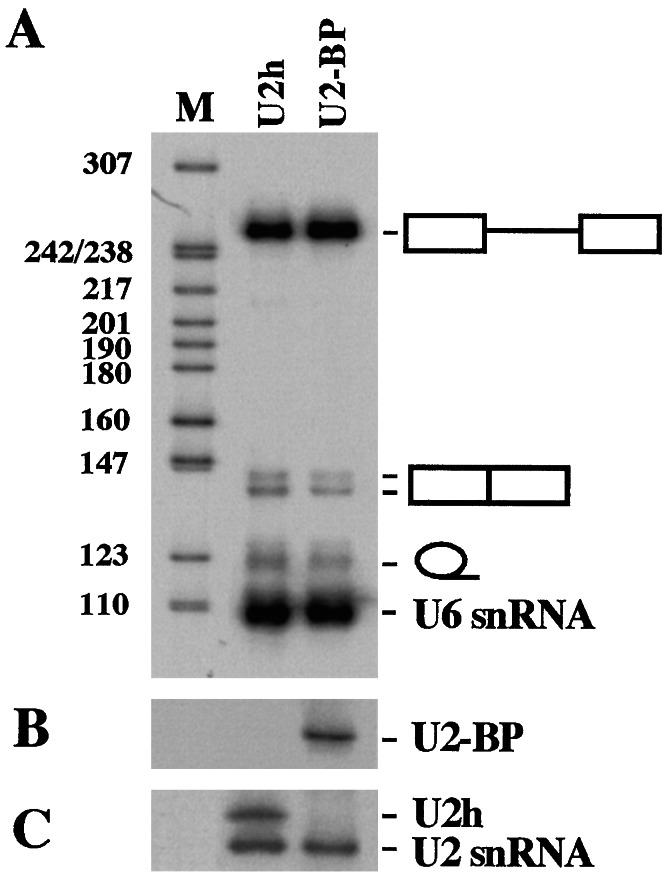
Chimeric U1 and U7 constructs were previously shown not to interfere with endogenous functions (17, 18). To analyze whether the excess of the modified form of U2 snRNA could interfere with the splicing machinery, we tested the accumulation of splicing products from a control pre-mRNA in the in vivo system of X. laevis oocytes. 32P-labeled AdML pre-mRNA was injected into oocyte nuclei previously injected with wild-type U2h (Fig. 3, lanes U2h) or U2-BP (Fig. 3, lanes U2-BP) genes. 32P-labeled U6 snRNA was coinjected as an internal control. Fig. 3A shows that no effect on the efficiency of splicing is observed when the U2 antisense construct is overexpressed in the nuclei of X. laevis oocytes in comparison with overexpression of wild-type U2h snRNA. Fig. 3B shows the expression of U2-BP snRNA, whereas Fig. 3C shows the expression of the control U2h snRNA. The U2h-specific expression is documented by the presence of a pre-U2 snRNA, which was shown to specifically accumulate in oocytes (26). From these data it can be concluded that the overexpression of U2-BP does not affect the overall efficiency of the splicing machinery.
Figure 3.
Effect on splicing of the U2-antisense construct. Oocytes were injected with wild-type U2h (lane U2h) or U2-BP (lane U2-BP) plasmids and subsequently with a 32P-labeled AdML pre-mRNA. RNA from nuclei of single oocytes was run on a 6% polyacrylamide/7 M urea gel and blotted. Direct autoradiography of the filter allows the visualization of 32P-labeled RNAs (A), whereas hybridizations with specific probes allow to analyze the expression of the injected plasmids (B and C). In B an antisense specific probe was used, whereas in C, a U2 snRNA probe was used. The U2h snRNA in oocytes mainly accumulates as a precursor few nucleotides longer (band U2h, see ref. 26). The different splicing products are indicated on the side. Lane M, pBR322 plasmid DNA, digested with MspI.
Analysis of the mRNA Produced in Response to the Antisense Treatment.
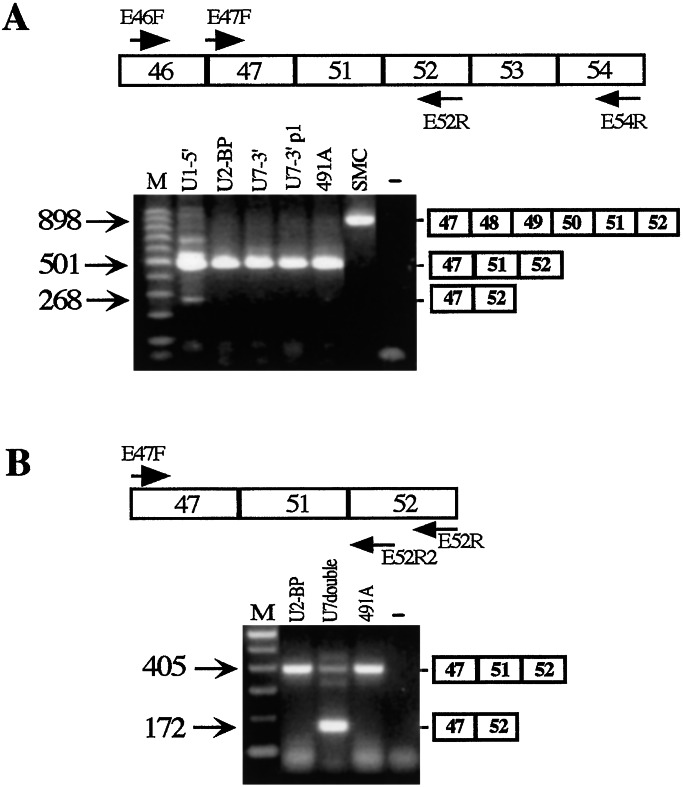
Mixed cell populations expressing different types of single chimeric constructs were analyzed by RT-PCR for the presence or absence of exon 51 in the mature mRNA. Fig. 4 shows the gel analysis of the RT-PCR products obtained with specific couples of oligos as specified in the top of each panel. Fig. 4A shows that among the first series of cells tested, only U1–5′ gave a faint band corresponding to the size of a skipped product. If compared with the amount of the unskipped product, it appears that less than 10% of the mRNA was deprived of exon 51. Interestingly, when the U7-double construct was analyzed, effective skipping was observed (Fig. 4B). In this case, more than 60% of the mRNA lacks exon 51. Because U7 double contains two antisense elements inside the same RNA molecule, we deduced that to have effective skipping, both splice junctions should be masked to the splicing machinery.
Figure 4.
Analysis of the effect of single antisense constructs on the splicing pattern of dystrophin mRNA. In the upper part of the panels, the oligos used for the nested RT-PCR reactions are indicated on the 491A dystrophin pre-mRNA. RT-PCR was performed with the external oligos; after 40 rounds of amplification, nested PCR was performed with the internal oligos. The amplification products from untreated (lanes 491A) and transduced (the specific antisense construct is indicated above each lane) cell lines were run on 2% agarose gels in parallel with a RNA-minus control (“−” lane) and with the products of amplification obtained on RNA from a wild-type skeletal muscle cell line (lane SMC in A). (A) The 501-nt-long amplification product corresponds to the unskipped 491A RNA, whereas the 268-nt-long fragment corresponds to the skipped mRNA. In lane SMC, the 898-nt-long amplification product has the size expected for a wild-type dystrophin mRNA. (B) The 405-nt-long amplification product corresponds to the unskipped product of the 491A RNA, whereas the 172-nt-long fragment corresponds to the skipped mRNA. A schematic representation of the unskipped and skipped products is shown on the side of the gels. Lanes M, 100-bp DNA ladder (Invitrogen).
To test whether this was the case, we made additional constructs in which couples of antisense constructs were cloned in tandem inside the pBabe retroviral vector (Fig. 5A). The expression of these double antisense constructs was tested in vivo, in puromycin-selected transduced cells, by Northern analysis. Fig. 5B shows that the head-to-tail cloning does not interfere with efficient expression of both chimeric RNAs.
Figure 5.
Analysis of the effect of double antisense constructs on the splicing pattern of dystrophin mRNA. (A) Schematic representation of pBabe puro retroviral constructs. The antisense-expressing cassettes were cloned head-to-tail in forward orientation in the 3′ long terminal repeat. Vector 5′/BP contains constructs U2-BP and U1–5′, whereas 5′/3′ contains U1–5′ and U7–3′p1. (B) Northern blot analysis was performed on 8 μg of total RNA extracted from 491A cells untransduced (lanes 491A) or transduced with the vectors indicated above the lanes. Hybridizations were carried out with 32P-labeled oligonucleotides indicated below. Lane M, pBR322 plasmid DNA, digested with MspI. (C) Nested RT-PCR was carried out as in Fig. 4B on RNA from control untransduced cells (lane 491A) and from cells transduced with the double constructs (lanes 5′/3′ and 5′/BP). Lane −, control amplification in the absence of RNA. Lane M, 100-bp DNA ladder (Invitrogen). A schematic representation of the unskipped and skipped products is shown on the side of the gel. (Lower) The sequence of the shorter RT-PCR products shows the precise skipping of exon 51. (D) Total proteins (100 μg) extracted from 491A cells untransduced (lanes 491A) or transduced with the vectors indicated above the lanes were run on a SDS/6% polyacrylamide gel in parallel with proteins extracted from wild-type skeletal muscle cells (lane SMC). After blotting the upper part of the gel, containing proteins of molecular mass higher than 200 kDa, was reacted with anti-dystrophin antibodies (Upper), whereas the lower part with anti-myosin antibodies (Lower). The asterisk indicates a degradative form of dystrophin as already reported (27).
DMD myoblasts carrying the double antisense constructs were then analyzed for exon 51 skipping by RT-PCR. Fig. 5C shows that both constructs 5′/3′ and 5′/BP produce the appearance of a band corresponding to the size of a skipped product (172 nucleotide long). Sequence analysis of the gel-purified band indicated that the skipped product is an exact fusion of exon 47 with exon 52. These data supported the previous hypothesis that both splice junctions should be masked by antisense sequences to have efficient skipping. In the case of 5′/3′ and 5′/BP construct, the amount of skipped product was in the order of 30–40%.
Rescue of Dystrophin Synthesis.
Immortalized 491A cells transduced with the double antisense constructs were analyzed for dystrophin synthesis by Western analysis (Fig. 5D Upper). In comparison to untreated DMD cells (lane 491A), those expressing the antisense constructs show rescue of dystrophin synthesis. The levels of dystrophin are fairly good if compared with those present in the control of skeletal muscle cells (lane SMC). In the latter sample the faster migration band (indicated by an asterisk) represents a specific degradation product of dystrophin as already reported (27). The lack of size difference observed between normal and induced dystrophin on the Western blot was expected, because removal of exons 48 to 51, which codes for 210 amino acids, would not significantly alter the migration of such a large (427-kDa) protein in a polyacrylamide gel. Fig. 5D Lower shows a control Western analysis on the same gel with myosin-specific antibodies.
Discussion
Many properties of RNA, if appropriately modified, can represent powerful tools for controlling gene expression, at the posttranscriptional level, in a sequence-specific way (17, 18, 28, 29). In human, this approach finds interesting applications in the gene therapy of inherited and acquired genetic disorders where one wants to repress or modify the expression of specific genes. One of the basic features of RNA is to function as an antisense molecule. The antisense strategy has been largely used in the past to interfere with translation of mRNAs (30) and, more recently, to interfere with nuclear processes, such as splicing (31–33). Even if such kind of control can be accomplished by the use of synthetic oligonucleotides (10–13), a significant drawback to this approach is caused by the fact that oligonucleotides would require periodic administrations. To circumvent this problem, over the last few years a lot of effort has been devoted to the production of several vectors able to express in vivo, in a stable fashion, large amounts of chimeric RNAs containing therapeutic sequences (17, 29, 33–35).
To obtain effective “therapeutic” RNA molecules in vivo, several important parameters must be ensured: not only they must be cloned under efficient promoters but, in addition, the RNA context in which the therapeutic RNA is embedded should provide stability and specific subcellular localization. The last point is a very crucial one because in the cell RNAs are sorted to specific cellular locations (nucleus, nucleolus, cytoplasm, etc.) and efficient activity of the therapeutic molecule can be obtained only if colocalization with the target is ensured (17, 36, 37).
Small cellular RNAs have been used as vectors because they are normally transcribed from strong promoters (polII or polIII) and because they allow the delivery of the chimeric constructs into specific subcellular compartments (17, 29, 33, 35, 36).
DMD is a genetic disease where the antisense strategy can find very powerful applications. Because in most of the cases deletions or point mutations in the huge dystrophin gene produce premature stop codons, it would be possible by preventing the inclusion of specific exons to produce a shorter but functional mRNA. This effect can be obtained through the expression of antisense molecules raised against specific splicing junctions. We have tested whether, in the case of a patient with the exons 48–50 deletion, it was possible to induce exon 51 skipping. This induction would produce an in-frame mRNA and eventually rescue the synthesis of a slightly shorter but presumably functional dystrophin. Interestingly, DMD deletions that by skipping of exon 51 would be corrected into in-frame mRNA have been calculated to represent almost 15% of all DMD (DMD–Leiden Database, http://www.dmd.nl).
The choice of the carrier RNAs was dictated by the possibility of colocalizing the antisense molecules with their target RNA. Two of the selected RNAs, U1 and U2, are known to participate in the splicing reaction, thus increasing the probability that the antisense molecule colocalizes with the dystrophin pre-mRNA. In addition, both selected RNAs recognize the pre-mRNA substrate by base pairing interaction. The third vector used consisted in the nucleoplasmic U7 snRNA (21). This RNA was previously used to express antisense molecules in vivo and shown to modify the splicing patter of a mutant β-globin pre-mRNA (18, 35).
The U1 and U2 snRNA promoters are polII-dependent and were shown to direct high levels of transcripts (17, 38). On the contrary, the U7 snRNA gene promoter is known to be less active (19, 38). For this reason the U7 constructs were also cloned under a U1 snRNA promoter. Nevertheless, from our analysis it appears that higher levels of chimeric molecules accumulate when U7 constructs are transcribed from their own promoter.
Analysis of cell lines derived from a dystrophic patient, immortalized with a SV40 large T, and stably transduced with retroviral vectors carrying the different constructs indicated that efficient skipping was obtained when two antisense molecules specific for the 5′ and 3′ splice junctions of exon 51 were coexpressed in the same cell. Lower effect was observed when single antisense molecules were used. In line with the idea that maximal effect could be obtained when both the 5′ and 3′ splice junctions are targeted with antisense molecules, we found that a U7 vector carrying a double antisense insert covering both the 5′ and 3′ splice sites was able to induce efficient skipping of exon 51.
In those cells where exon 51 skipping was observed, rescue of dystrophin synthesis was obtained indicating that the mRNA was indeed converted into a functional form.
Further work on the extension of this strategy on animal systems is needed to prove whether this approach can be used for permanent correction of the mutation in vivo.
Acknowledgments
We are particularly grateful to Daniel Schumperli, Daniel Sutter, and Alan Weiner for providing plasmids and for helpful suggestions. We thank all of the members of the Bozzoni laboratory for helpful discussion. We thank M. Arceci and G. Ricci for skilful technical help. This work was partially supported by grants from Telethon–Italy (Grants 1195 and 1322), Ministero dell'Universitá e della Ricerca Scientifica e Tecnologica (PRIN 40% and “Centro di eccellenza BEMM”), and Consiglio Nazionale delle Ricerche (“Target Project on Biotechnology” and “Tecnologie di base della postgenomica”).
Abbreviations
- DMD
Duchenne muscular dystrophy
- snRNA
small nuclear RNA
References
- 1.Koenig M, Beggs A H, Moyer M, Scherpf S, Heindrich K, Bettecken T, Meng G, Muller C R, Lindlof M, Kaariainen H, et al. Am J Hum Genet. 1989;45:498–506. [PMC free article] [PubMed] [Google Scholar]
- 2.Hodgson S V, Bobrow M. Br Med Bull. 1989;45:719–744. doi: 10.1093/oxfordjournals.bmb.a072354. [DOI] [PubMed] [Google Scholar]
- 3.Zatz M, Lange K, Spence M A. Lancet. 1977;1:759. doi: 10.1016/s0140-6736(77)92211-5. [DOI] [PubMed] [Google Scholar]
- 4.Gussoni E, Blau H M, Kunkel L M. Nat Med. 1997;3:970–977. doi: 10.1038/nm0997-970. [DOI] [PubMed] [Google Scholar]
- 5.Partridge T A, Morgan J E, Coulton G R, Hoffman E P, Kunkel L M. Nature (London) 1989;337:176–179. doi: 10.1038/337176a0. [DOI] [PubMed] [Google Scholar]
- 6.Wang B, Li J, Xiao X. Proc Natl Acad Sci USA. 2000;97:13714–13719. doi: 10.1073/pnas.240335297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campeau P, Chapdelaine P, Seigneurin-Venin S, Massie B, Tremblay J P. Gene Ther. 2001;8:1387–1394. doi: 10.1038/sj.gt.3301532. [DOI] [PubMed] [Google Scholar]
- 8.Cho W K, Ebihara S, Nalbantoglu J, Gilbert R, Massie B, Holland P, Karpati G, Petrof B J. Hum Gene Ther. 2000;11:701–714. doi: 10.1089/10430340050015608. [DOI] [PubMed] [Google Scholar]
- 9.Chen H H, Mack L M, Kelly R, Ontell M, Kochanek S, Clemens P R. Proc Natl Acad Sci USA. 1997;94:1645–1650. doi: 10.1073/pnas.94.5.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Deutekom J C, Bremmer-Bout M, Janson A A, Ginjaar I B, Baas F, den Dunnen J T, van Ommen G J. Hum Mol Genet. 2001;10:1547–1554. doi: 10.1093/hmg/10.15.1547. [DOI] [PubMed] [Google Scholar]
- 11.Mann C J, Honeyman K, Cheng A J, Ly T, Lloyd F, Fletcher S, Morgan J E, Partridge T A, Wilton S D. Proc Natl Acad Sci USA. 2001;98:42–47. doi: 10.1073/pnas.011408598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunckley M G, Manoharan M, Villiet P, Eperon I C, Dickson G. Hum Mol Genet. 1998;7:1083–1090. doi: 10.1093/hmg/7.7.1083. [DOI] [PubMed] [Google Scholar]
- 13.Wilton S D, Lloyd F, Carville K, Fletcher S, Honeyman K, Agrawal S, Kole R. Neuromuscul Disord. 1999;9:330–338. doi: 10.1016/s0960-8966(99)00010-3. [DOI] [PubMed] [Google Scholar]
- 14.Morgestern J P, Land H. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fragapane P, Caffarelli E, Lener M, Prislei S, Santoro B, Bozzoni I. Mol Cell Biol. 1992;12:1117–1125. doi: 10.1128/mcb.12.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhuang Y, Weiner A M. Cell. 1986;46:827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- 17.Michienzi A, Prislei S, Bozzoni I. Proc Natl Acad Sci USA. 1996;93:7219–7224. doi: 10.1073/pnas.93.14.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorman L, Suter D, Emerick V, Schumperli D, Kole R. Proc Natl Acad Sci USA. 1998;95:4929–4934. doi: 10.1073/pnas.95.9.4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grimm C, Stefanovic B, Schumperli D. EMBO J. 1993;12:1229–1238. doi: 10.1002/j.1460-2075.1993.tb05764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galli G, Hofstetter H, Stunnenberg H G, Birnstiel M L. Cell. 1983;34:823–828. doi: 10.1016/0092-8674(83)90539-1. [DOI] [PubMed] [Google Scholar]
- 21.Bond U M, Yario T A, Steitz J A. Genes Dev. 1991;5:1709–1722. doi: 10.1101/gad.5.9.1709. [DOI] [PubMed] [Google Scholar]
- 22.Spycher C, Streit A, Stefanovic B, Albrecht D, Koning T H, Schumperli D. Nucleic Acids Res. 1994;22:4023–4030. doi: 10.1093/nar/22.20.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Datta B, Weiner A M. Nature (London) 1991;352:821–824. doi: 10.1038/352821a0. [DOI] [PubMed] [Google Scholar]
- 24.Sun J S, Manley J L. Genes Dev. 1995;9:843–854. doi: 10.1101/gad.9.7.843. [DOI] [PubMed] [Google Scholar]
- 25.Berghella L, De Angelis L, Coletta M, Berarducci B, Sonnino C, Salvatori G, Anthonissen C, Cooper R, Butler-Browne G S, Mouly V, et al. Hum Gene Ther. 1999;10:1607–1617. doi: 10.1089/10430349950017617. [DOI] [PubMed] [Google Scholar]
- 26.You C-H, Ares M, Jr, Weiner A M. Cell. 1985;42:193–202. doi: 10.1016/s0092-8674(85)80115-x. [DOI] [PubMed] [Google Scholar]
- 27.Anderson L V B, Davison K. Am J Pathol. 1999;154:1017–1022. doi: 10.1016/S0002-9440(10)65354-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cech T R. Biochem Int. 1989;18:7–14. [PubMed] [Google Scholar]
- 29.Buonomo S B, Michienzi A, De Angelis F G, Bozzoni I. RNA. 1999;5:993–1002. doi: 10.1017/s1355838299990064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crooke S T. Antisense Nucleic Acid Drug Dev. 1998;8:133–134. doi: 10.1089/oli.1.1998.8.133. [DOI] [PubMed] [Google Scholar]
- 31.Kole R, Sazani P. Curr Opin Mol Ther. 2001;3:229–234. [PubMed] [Google Scholar]
- 32.Sierakowska H, Sambade M J, Agrawal S, Kole R. Proc Natl Acad Sci USA. 1996;93:12840–12844. doi: 10.1073/pnas.93.23.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorman L, Mercatante D R, Kole R. J Biol Chem. 2000;275:35914–35919. doi: 10.1074/jbc.M006259200. [DOI] [PubMed] [Google Scholar]
- 34.Michienzi A, Cagnon L, Bahner I, Rossi J J. Proc Natl Acad Sci USA. 2000;97:8955–8960. doi: 10.1073/pnas.97.16.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suter D, Tomasini R, Reber U, Gorman L, Kole R, Schumperli D. Hum Mol Genet. 1999;8:2415–2423. doi: 10.1093/hmg/8.13.2415. [DOI] [PubMed] [Google Scholar]
- 36.Bertrand E, Castanotto D, Zhou C, Carbonnelle C, Lee N S, Good P, Chatterjee S, Grange T, Pictet R, Kohn D, Engelke D, Rossi J J. RNA. 1997;3:75–88. [PMC free article] [PubMed] [Google Scholar]
- 37.Sullenger B A, Cech T R. Science. 1993;262:1566–1569. doi: 10.1126/science.8248806. [DOI] [PubMed] [Google Scholar]
- 38.Birnstiel M L, Schaufele F. In: Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles. Birnstiel M L, editor. Berlin: Springer; 1988. pp. 155–182. [Google Scholar]