Abstract
HIV-1 is cytopathic for CD4+ T lymphocytes in vitro and this property of HIV-1 is generally considered to account for some of its in vivo cytopathogenicity. Thus, the extent of lymphocyte depletion correlates with the level of viremia whereas low levels of viral replication are typically associated with stable lymphocyte levels and asymptomatic infection such as is observed in non-progressors. Here, we describe a non-progressor who did not fit this general pattern in that CD4+ T lymphocyte homeostasis was maintained in the face of high-level viral replication. Biological viral isolates from this patient replicated in primary lymphocytes without inducing cytopathicity. Because this phenotype is reminiscent of Vpr-deleted viruses, we examined the contribution of the Vpr gene to the viral phenotype. Vpr alleles derived from this patient contained both premature stop codons and an unusual Q3R polymorphism. Insertion of patient-derived Vpr alleles or a Q3R substitution into a cytopathic HIV-1 clone resulted in a marked impairment of cytopathicity without affecting viral replication efficiency. The effect of Vpr on cytopathicity was unrelated to reported activities of Vpr including virion association, interaction with uracil DNA glycosylase, G2 arrest, or enhancement of macrophage infection but correlated with the ability of Vpr to induce host cell apoptosis. This study suggests the presence of a determinant of in vivo cytopathogenicity within HIV-1 Vpr and further indicates that viral replication can be uncoupled from cytopathicity in vitro and in vivo.
Keywords: non-progressive infection‖accessory proteins‖apoptosis‖pathogenesis
HIV-1 infection of humans is characterized by progressive destruction of CD4+ T lymphocytes. The number of circulating CD4+ T lymphocytes is inversely correlated with the level of viral replication, and persistent high viral loads in plasma predict a rapid disease onset (1). At the other end of the spectrum, individuals with low or undetectable plasma viremia exhibit a non-progressive infection (1). Although HIV-1 is directly cytopathic for CD4+ T cells in vitro, the largely debated issue is whether lymphocyte depletion in vivo is a consequence of viral cytopathicity, lymphocyte redistribution between the circulation and lymphoid organs, activation-induced death of uninfected cells, or impaired hematopoieis (2).
The mechanism by which HIV-1 effects cytopathicity in infected CD4+ T lymphocytes in vitro remains poorly characterized. Acute cytopathicity is reflected by syncytium formation and syncytium-independent single cell killing, both of which can be mediated by the fusogenic characteristics of the viral envelope glycoprotein (3–5). Envelope glycoprotein also mediates virus attachment to receptor and coreceptor molecules on the cell surface, and the ability of HIV-1 strains to induce cytopathic effects in vitro reflects syncytium-inducing and coreceptor usage characteristics of the envelope glycoprotein. Highly cytopathic variants use CXCR4 as an entry cofactor and induce syncytia in T cell lines (SI viruses) whereas less cytopathic variants use CCR5 and are non-syncytium-inducing (NSI viruses) (6). These in vitro characteristics appear to have prognostic significance in that the presence of SI variants generally reflects a faster course of disease (7, 8). Much effort has focused on the involvement of apoptosis in HIV-1-mediated cell death (reviewed in ref. 9). Apoptotic and anti-apoptotic effects in vitro have been described for a number of viral gene products including Env, Tat, Nef, and Vpr. However, those effects that are operative in vivo to impair CD4+ T lymphocyte homeostasis are unclear.
Whereas the magnitude of viral replication, as measured by plasma viral load, is predictive of the rate of disease progression, asymptomatic infection is reflected by low to undetectable levels of viral replication and CD4+ T lymphocyte homeostasis (10, 11). This non-progressive infection is the result of host genetic factors such as coreceptor polymorphisms (12), which may limit the permissiveness of the host to viral replication, as well as viral factors such as accessory gene defects, which may impact viral replication capacity (13–15). The object of this study, patient P1016, is an exception to the non-progressor phenotype in that CD4 lymphocyte levels were maintained at normal levels despite high levels of plasma viral RNA. Thus, viral replication and lymphocyte depletion appeared to have been uncoupled in this patient. In an attempt to identify the basis for this unusual phenotype, we focused on viral characteristics that would similarly uncouple viral replication from viral cytopathicity in vitro.
Materials and Methods
Patient Characteristics and Virus Isolation.
P-1016 is a child with vertical HIV-1 infection. Antiretroviral therapy (Nevirapine monotherapy) was initiated at 4 yr of age due to persistently high plasma viral loads (up to 330,000 HIV-1 RNA copies/ml). Despite the subsequent addition of zidovudine and didanosine to the regimen, plasma HIV-1 RNA levels remained elevated > the 95th percentile through at least 5 yr of age. This patient has remained asymptomatic with normal CD4 counts relative to age-matched controls through 15 years of age, despite the high early plasma HIV-1 RNA. Normal median absolute CD4 T cell numbers for individuals aged 2–5 years and 7–17 years are 1,800 and 800, respectively (16, 17). Absolute CD4 numbers for patient P-1016 were within the normal range for age-matched controls despite intermittent episodes of viremia in excess of 1.105 RNA copies per ml of plasma.
Analysis of Viral Cytopathicity.
Donor peripheral blood mononuclear cells (PBMCs) were depleted of CD8 cells by using anti-CD8 antibody-coated magnetic beads (Dynal, Lake Success, NY) according to the manufacturer's protocol. CD8-depleted T cells were stimulated overnight with phytohemagglutinin and then cultured at 1.106 cells per ml in RPMI 1640 supplemented with 10% FCS and recombinant human interleukin 2 (10 units/ml; Boehringer-Mannheim). To eliminate the contribution of syncytia to viral cytopathic effects, cultures were maintained in the continuous presence of azide-free anti-CD4 antibody (BioSource International, Camarillo, CA) at 100 ng/ml after virus infection. Cell viability at various intervals postinfection was evaluated by trypan blue dye exclusion.
Analysis of Cellular Apoptosis.
Apoptotic activity of Vpr was evaluated in HeLa cells transfected with infectious HIV-1 molecular clones (HIV-1 LAI) carrying LAI Vpr, patient-derived Vpr alleles, or mutants thereof. Briefly, 3.104 HeLa cells were seeded in 24-well plates, and, 24 hr later, cells were transfected with 3–4.5 μg of proviral DNA by calcium phosphate/DNA coprecipitation. At 16 hr posttransfection, the extent of the host cell apoptosis was determined by measuring cytoplasmic histone-associated DNA fragments (mono- and oligonucleosomes) by ELISA (Cell Death Detection ELISAplus; Roche Molecular Biochemicals). Actinomycin D (0.1 μg/ml) was used as a positive control for indication of apoptosis. Vpr apoptotic activity was further examined in an independent assay of annexin V staining (Apopnexin Detection Kit, Intergen, Purchase, NY). After transfection of HeLa cells with expression plasmids (pME379, provided by M. Emerman, Fred Hutchinson Cancer Research Center, Seattle) containing patient-derived Vpr allele (clone I) with and without the Q3R polymorphism, cells were visualized by fluorescence microscopy and compared with nontransfected cells, cells transfected with nonspecific DNA, and cells treated with actinomycin D.
Cells, Viruses, and Reagents.
HIV-1 LAI/ADA contains the envelope V3 region of the monocytotropic HIV-1 ADA isolate and has been engineered to contain HpaI and XmaI restriction sites flanking the Vpr gene (M. Sharkey, unpublished results). Plasma-derived Vpr alleles were cloned between XmaI and HpaI sites in place of the parental HIV-1 LAI Vpr. Vpr point mutants in HIV-1 LAI were generated by PCR mutagenesis and inserted into the parental clone via NdeI sites. For lymphocyte infections, the Vpr alleles were inserted within corresponding regions of HIV-1 LAI (SI, X4) or HIV-1 LAI/ADA (NSI, R5). Macrophages were prepared from elutriated monocytes as described (18). HeLa cells and 293T were maintained in DMEM supplemented with 10% FCS. For generation of viral stocks, 293T cells were transfected with 20–30 μg proviral DNA by calcium phosphate/DNA coprecipitation. For macrophage infections, viruses were amplified in phytohemagglutinin-activated human peripheral blood lymphocytes. Infected cell frequency was determined by indirect immunofluorescence by using monoclonal anti-Gag p24 antibody (Kal-1; DAKO) and a secondary FITC-conjugated anti-mouse antibody (Jackson ImmunoResearch). HIV-1 gag p24 protein was quantified by ELISA (Coulter).
Analysis of Vpr Encapsidation.
HIV-1 LAI/ADA virions containing wild-type and patient-derived Vpr alleles were obtained from culture supernatants of transfected 293T cells. The relative levels of Vpr within cell lysates and purified virions were determined by Western blotting. Blots were probed in parallel with an antibody to gag MA, a major structural virion protein.
Analysis of Vpr/Uracil DNA Glycosylase (UDG) Association.
The interaction of Vpr with UDG was determined in a yeast-2-hybrid system. Briefly, the C-terminal 221 aa of human uracil N-glycosylase (UDG Δ84; ref. 19) was fused to the C terminus of the activation domain of GAL4. LAI Vpr or patient-derived Vpr alleles (clones I-IV) were inserted into the plasmid pAS2-1 as C-terminal fusions of the DNA binding domain of GAL4. The yeast reporter strain YRG2 (Stratagene), containing the GAL4 inducible reporter genes β-galactosidase (β-gal) and His-3, was cotransformed with the plasmid pACT2 (Δ84UDG) and pAS2-1 (Vpr). Cotransformants were selected by plating yeast on media deficient in leucine and tryptophan. β-Gal activity was measured in a liquid assay by using chlorophenol red β−d-galactopyranoside (CPRG) as a substrate. UDG interaction on the basis of β-gal activity was scored as follows: equivalent to LAI Vpr (++), in excess of LAI Vpr (+++), or no interaction (−) (see Fig. 3D).
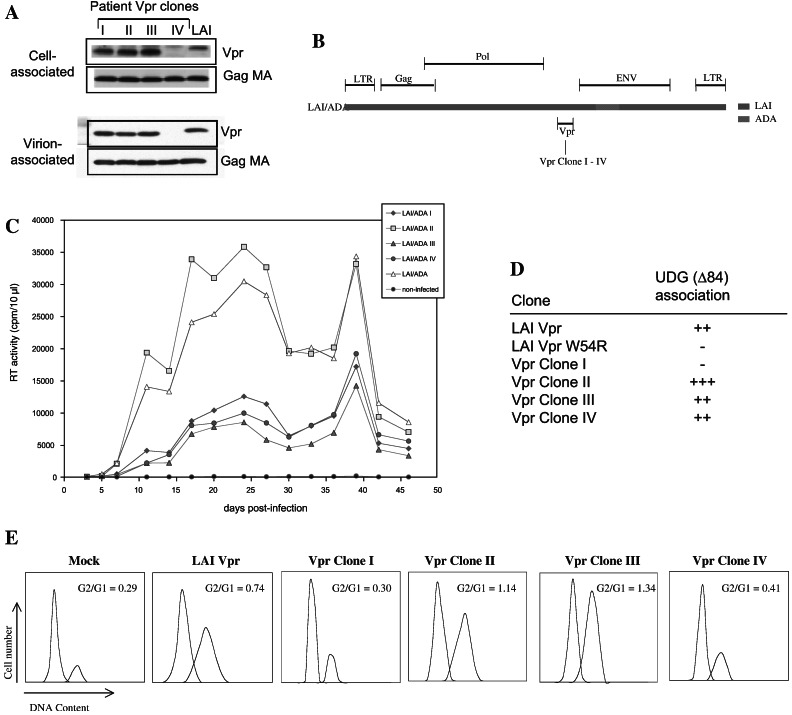
Figure 3.
In vitro activities of patient-derived Vpr alleles. (A) Analysis of relative levels of cell-associated and virion-associated Vpr proteins. (B) Schematic of HIV-1 LAI/ADA, an R5-tropic variant of HIV-1 LAI that contains envelope sequences spanning the V3 region of HIV-1ADA. (C) Replication profiles of LAI/ADA variants containing LAI Vpr and patient Vpr alleles in monocyte-derived macrophages. (D) Interaction of Vpr with uracil DNA glycosylase. (E) Induction of G2 arrest. G1 and G2 profiles for 293T cells expressing wild-type and patient-derived Vpr clones was determined specifically for productively infected cells.
Analysis of G2 Arrest.
The ability of patient-derived Vpr alleles to induce G2 arrest was determined as described (20). Briefly, LAI Vpr and patient-derived Vpr alleles were inserted into pEQ222 under control of the HIV-1 LTR. 293T cells were cotransfected with the Vpr plasmids and with plasmids expressing HIV-1 Tat and a nucleus-localized green fluorescent protein (GFP) under control of the HIV-1 long terminal repeat (LTR). Forty-eight hours after transfection, the cells were washed, fixed, stained with propidium iodide, and analyzed by cell sorting. GFP expression was determined by fluorescence emission at 530 nm after excitation at 488 nm. DNA profiles were analyzed by the multicycle AV program (Phoenix Flow Systems, San Diego).
Characterization of Vpr Alleles in Plasma.
Vpr genes were directly amplified from plasma viral RNA as described (21). To analyze the frequency of truncated Vpr alleles in plasma virions, recombinant PCR-script clones containing amplified Vpr alleles were analyzed by oligonucleotide probe-specific hybridization (21). Briefly, XL-1 Blue MRF-Kan cells were transformed with recombinant PCR clones and transferred to nylon membranes (Magna Lift; Micron Separations, Westboro, MA) and disrupted after successive treatments with 10% SDS (3 min), 0.5 M NaOH/1.5 M NaCl (5 min), 0.5 M Tris⋅HCl, pH 8.0/1.5 M NaCl (5 min), and 2× SSPE [standard saline phosphate/EDTA (0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA)] for 5 min. Nucleic acids were bound to membranes by using UV radiation (Strata Linker; Stratagene) and hybridized with 5′ [γ-32P]ATP end-labeled oligonucleotide probes specific for truncated (5′-c5674 cca cag ctt agg gta at) and open Vpr genes (5′-c5674 cat agc tta ggg caat). Filters were hybridized in a solution containing 5× SSC, 1× Denhardt's reagent (0.02% polyvinylpyrrolidone/0.02% Ficoll/0.02% BSA), 0.1% SDS for 16 h at 37°C. Hybridized filters were washed in 3 M tetramethylammonium chloride, 50 mM Tris⋅HCl (pH 8.0), and 0.2% SDS for 15 min at room temperature and then at 52°C for 1 h. Membranes were then washed twice in a solution of 2× SSC, 0.1% SDS for 10 min at room temperature. Hybridized colonies were visualized on a molecular PhosphorImager (Molecular Dynamics).
Results
Vpr Defects in a Viremic Long-Term Non-Progressor.
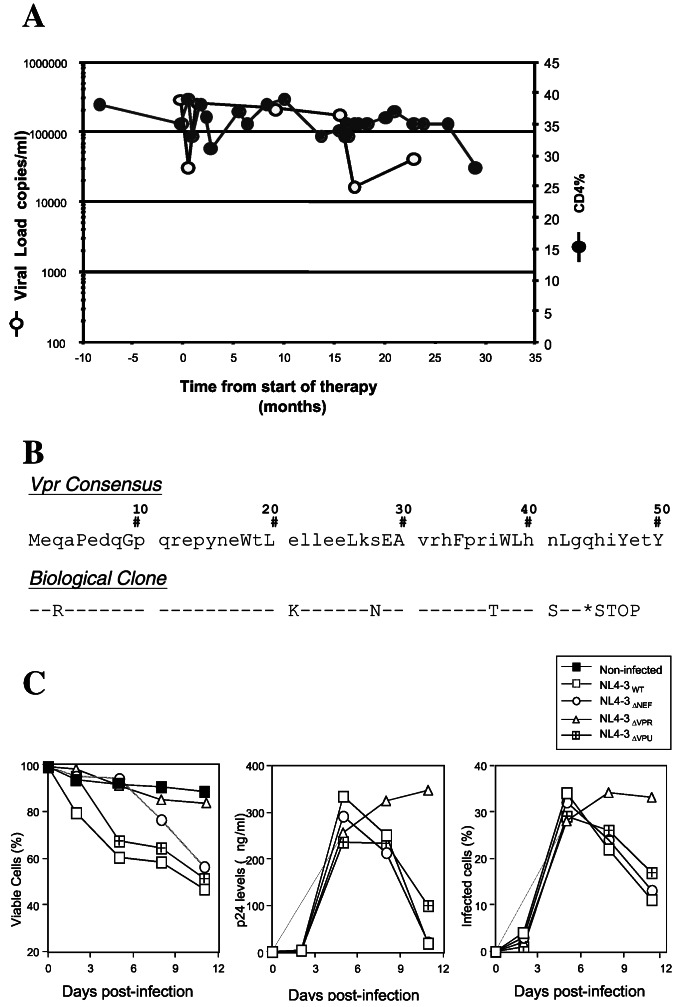
Biological clones were established from P1016 immediately before initiation of Nevirapine monotherapy (Fig. 1A). These biological virus isolates had an unusual phenotype in that they replicated within primary lymphocyte cultures without inducing cytopathicity that is characteristic of HIV-1 replication (not shown). This phenotype is reminiscent of viruses that emerge from long-term in vitro propagation and that develop spontaneous inactivating mutations in the Vpr gene (20, 22). Sequencing of the Vpr gene in these biological isolates revealed the presence of a premature stop codon that resulted in the production of a truncated Vpr protein of 43 aa (Fig. 1B). To confirm whether a defective vpr allele could uncouple viral replication and cytopathicity in vitro, the effects of a Vpr deletion on the phenotype of a prototypic cytopathic HIV-1 isolate (NL4-3) was compared with the phenotype of wild-type NL4-3 and NL4-3 variants lacking other accessory proteins such as Nef and Vpu. To exclude the formation of syncytia, which is a major determinant of viral cytopathicity in vitro, cells were infected and then cultured in the presence of a CD4 antibody. Because CD4 antibody blocks syncytium formation, any observed cytopathicity would be the result of single cell killing. The extent of viral replication both in terms of the frequency of infected cells and production of Gag p24 was equivalent in wild-type, ΔNef- and ΔVpr-infected cultures whereas there was slightly less extracellular p24 in cultures infected with the Vpu mutant. In contrast to the progressive reduction in cell viability in cultures infected with wild-type, ΔNef and ΔVpu viruses, cell viability in ΔVpr-infected cultures paralleled that of uninfected cultures (Fig. 1C). These results suggest that, in single cell-killing assays, inactivation of Vpr is sufficient to uncouple viral replication and cytopathicity.
Figure 1.
Patient characteristics and impact of Vpr mutations on viral cytopathicity. (A) Plasma viral RNA and CD4 lymphocyte profiles in patient P1016 with asymptomatic infection. Time intervals are shown relative to initiation of Nevirapine monotherapy. (B) Vpr truncation in virus isolates established from P1016 immediately before initiation of therapy. (C) In vitro cytopathic phenotype of wild-type and accessory gene deletion mutants of HIV-1.
Characteristics of Patient-Derived Vpr Alleles.
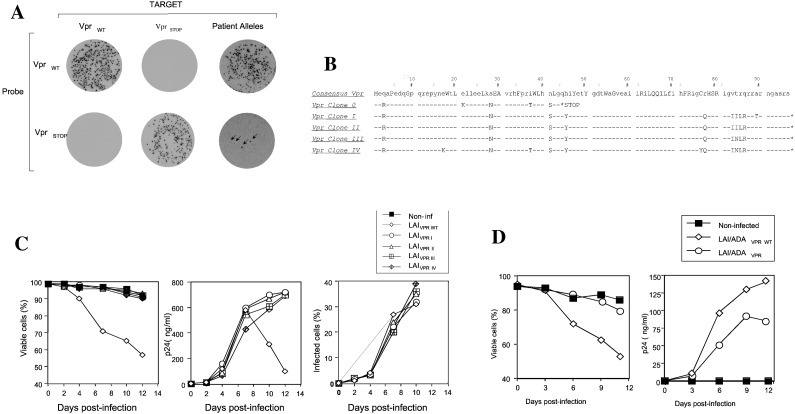
Although biological clones established from the viremic non-progressor contained a truncated vpr gene, such truncations are frequently generated by in vitro passage of HIV-1 (20, 22). We initially determined whether the premature stop codon identified in patient biological isolates was representative of the viral population in plasma. Vpr alleles were amplified from plasma viral RNA by PCR. Over 200 recombinant Vpr PCR clones were screened by probe-specific colony hybridization (21) using a probe that specifically recognizes the premature termination sequence (Fig. 2A). Only four (indicated by arrows) of the recombinant Vpr clones hybridized to the premature stop-specific probe indicating that viruses with these truncated Vpr alleles represented a minor population (≈1%) of plasma virions. To determine the presence of other mutations or polymorphisms that would impact the Vpr gene in this patient, over 20 nontruncated Vpr clones were selected and sequenced. Four representative Vpr alleles (Fig. 2B) were inserted into corresponding regions of the cytopathic infectious molecular clone HIV-1 LAI/ADA. The parental HIV-1 LAI variant caused rapid single cell killing in primary lymphocyte cultures (Fig. 2C). In contrast, the chimeric LAI variants containing patient-derived vpr alleles replicated without inducing cell death (Fig. 2C). The impact of Vpr on cytopathicity was independent of SI and coreceptor usage characteristics of the virus because insertion of one of the patient vpr alleles (clone I) into an NSI/R5 virus (LAI/ADA) also impaired viral cytopathicity (Fig. 2D). Thus, Vpr alleles derived from this non-progressor appeared to lack a cytopathic determinant that is present in prototypic (NL4-3, LAI) Vpr proteins.
Figure 2.
Cytopathic activity of Vpr alleles derived from plasma viral RNA. (A) Frequency of Vpr alleles containing premature stop codons as assessed by oligonucleotide probe-specific hybridization. Bacterial colonies transformed with wild-type, Vprstop and patient-derived Vpr alleles were hybridized with probes recognizing wild-type or truncated Vpr sequences. Hybridization conditions were adjusted to promote specific binding of each probe to its cognate sequence. One of four clones hybridizing specifically to the Vpr stop probe (clone 0) and four hybridizing specifically to the wild-type Vpr probe (clones I-IV) were sequenced (B). Consensus Vpr sequences were derived from 101 clade B Vpr sequences listed in the Los Alamos HIV-1 Sequence Database. (C and D) Replication and cytopathicity of X4-tropic (C) and R5-tropic (D) HIV-1 LAI variants containing wild-type and patient-derived Vpr alleles.
A number of in vitro activities have been described for Vpr, including the induction of G2 arrest, enhancement of infection of nondividing macrophages, and interaction with the DNA repair enzyme UDG (for review, see ref. 23). Vpr is encapsidated within virions through a specific interaction with the p6 domain of the p55 gag precursor, and virion incorporation is considered to be important for the role of Vpr in postentry events in viral replication, such as nuclear uptake of viral cDNA. We examined whether patient vpr alleles exhibited any gross defects, with regards to the reported activities of Vpr, that may account for the impaired cytopathicity of these vpr alleles. Vpr clones I, II, and III were efficiently expressed in mammalian cells. Clone IV was expressed at barely detectable levels (Fig. 3A). This finding is most likely a result of protein instability because of loss of a cysteine at amino acid 76 (Fig. 2B). Because clone IV was unstable, it served as a negative control for the analysis of Vpr-associated activities in mammalian cells. Vpr protein derived from clones I, II, and III was efficiently packaged within virions whereas, as expected, Vpr clone IV was not detected (Fig. 3A). To examine activities of the patient-derived vpr alleles with regard to macrophage infection, the alleles were inserted into the corresponding region of a macrophage-tropic infectious molecular clone HIV-1LAI/ADA (Fig. 3B). Because clone IV is unstable and defective for virion incorporation, it is incapable of facilitating macrophage infection. Replication of the chimeric viruses was compared with the parental HIV-1LAI/ADA clone. Replication of the LAI/ADA variant containing Vpr clone II was comparable to that of parental LAI/ADA in monocyte-derived macrophages (Fig. 3C). Replication of LAI/ADA variants containing Vpr clones I and III was impaired to the same extent as clone IV (Fig. 3C). Nevertheless, because all four clones similarly impaired viral cytopathicity, we conclude that there is no correlation between Vpr's effect on viral cytopathicity and virion encapsidation or macrophage infection. We next used the yeast 2-hybrid system to examine the association of UDG with the patient-derived vpr alleles. In this system, all patient Vpr clones, including clone IV, were efficiently expressed. Positive interactions were observed between UDG and patient Vpr alleles II, III, and IV, as well as the positive control, LAI Vpr (Fig. 3D). No interaction was observed between UDG and patient Vpr allele clone I or a Vpr W54R mutant shown previously to impair interaction between UDG and Vpr (19). To determine whether these patient-derived alleles were competent for G2 arrest, cell cycle profiles of 293T cells transfected with plasmids expressing HIV-1 LAI Vpr or patient-derived vpr alleles were examined. Two patient alleles (clones II and III) induced efficient G2 arrest similar to that of LAI Vpr whereas the cell cycle profiles of cells expressing clones I and IV were not significantly different from those of mock-transfected cells (Fig. 3E). Thus, the G2 arrest activity of Vpr did not correlate with the effects of Vpr on cytopathicity.
A Vpr Q3R Polymorphism Affects the Cytopathic and Apoptotic Activity of Vpr.
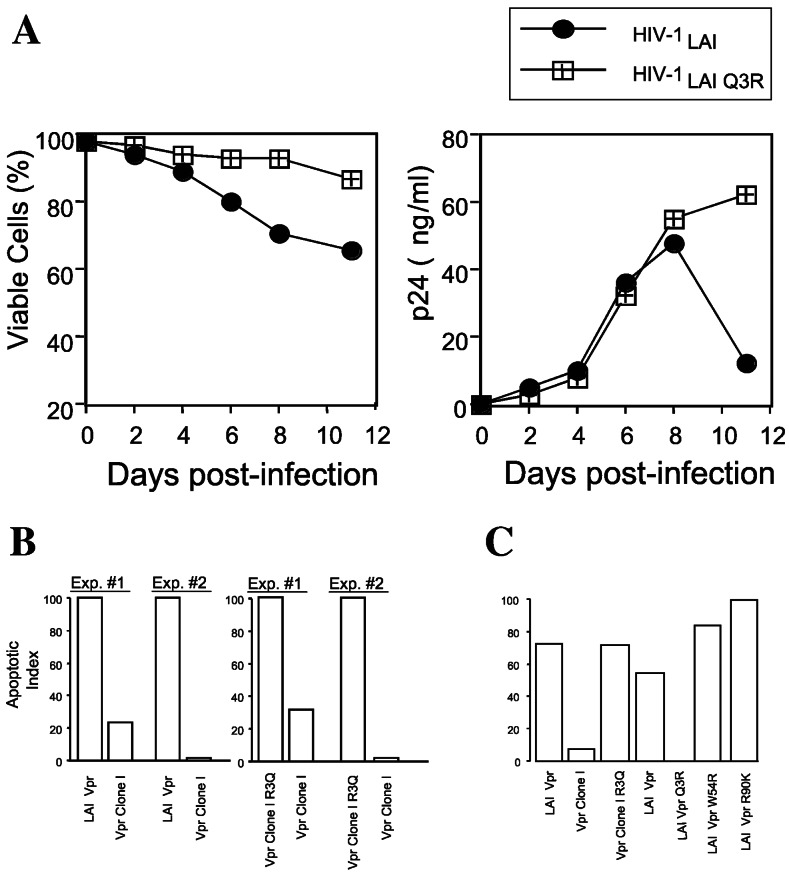
Patient-derived vpr alleles contained a highly unusual Q3R polymorphism at the N terminus of Vpr (Fig. 2B). In addition, multiple plasma viral RNA specimens obtained from P1016 at various intervals before initiation of therapy were sequenced and uniformly contained the Q3R polymorphism. Of 101 clade B Vpr sequences in the Los Alamos sequence database, only 2 contained polymorphisms at this amino acid. In addition, the Q3R mutation was seen only in one HIV-1 type N and one type O sequence in the database. By comparison, other non-consensus polymorphisms in patient-derived Vpr clones (for example, N28S, N41S, and H45Y) were common in Vpr alleles contained in the Los Alamos database. To determine whether the Q3R polymorphism was necessary and sufficient to recapitulate the noncytopathic phenotype, the single cell-killing characteristics of a cytopathic HIV-1 LAI variant and an HIV-1 LAI variant containing a Q3R polymorphism in Vpr were examined in primary lymphocytes. Whereas replication of the parental LAI clone was accompanied by cytopathicity, cultures infected with the LAI Q3R variant did not reveal any cytopathic effect (Fig. 4A), and cell viability paralleled that of uninfected cultures (not shown). Studies have demonstrated that the HIV-1-induced cytopathic effect correlates with the extent of host-cell apoptosis and that Vpr may contribute to the apoptotic effect (for review, see ref. 9). Furthermore, HIV-1 Vpr has been shown to induce the apoptosis of human neuronal cells after expression from a lentivirus vector (24). Therefore, we examined whether the differential cytopathic activity of viruses containing wild-type and Q3R vpr alleles reflected differential apoptotic activities in HeLa cells. Whereas cells expressing an LAI Vpr exhibited extensive apoptosis (DNA fragmentation), significantly less apoptosis was evident in cells expressing the noncytopathic vpr allele (Fig. 4B). Reversion of the Q3R polymorphism to the Vpr consensus sequence (R3Q) in clone I increased its apoptotic activity (Fig. 4B). Apoptotic activity was additionally evaluated by cell surface annexin V staining, an independent indicator of apoptosis. Cells transfected with Vpr clone I harboring a revertant (R3Q) clone I exhibited a pattern of annexin V staining that was similar to that of cells treated with actinomycin D whereas the level of annexin V staining in cells transfected with the Q3R clone was similar to that of nontransfected cells or cells transfected with a nonspecific DNA (not shown). Introduction of W54R and R90K substitutions, shown previously to influence UDG association and cell cycle arrest properties of Vpr, respectively, did not impair apoptotic activity (Fig. 4C.). The ability of the Q3R substitution to impair apoptotic activity of a wild-type Vpr and the ability of an R3Q reversion to restore apoptotic activity to a noncytopathic patient Vpr allele suggest that this polymorphism is sufficient to account for the noncytopathic characteristics of the patient-derived Vpr clones.
Figure 4.
Influence of the Q3R polymorphism on cytopathic and apoptotic activities of Vpr. (A) Cell viability and viral replication in CD8 depleted CD4+ T lymphocytes infected with wild-type HIV-1 LAI and an LAI variant containing a Q3R Vpr mutation. (B) Apoptotic activity of HIV-1 LAI/ADA variants containing LAI Vpr or a patient-derived Vpr allele (clone I). (C) Effect of amino acid substitutions on apoptotic activity of Vpr. The indicated amino acid substitutions were inserted within either LAI Vpr or a patient-derived vpr allele (clone I). The Q3R polymorphism in the patient-derived Vpr allele (clone I) was mutated to a Q3 consensus sequence (clone I R3Q). W54R and R90K substitutions impair UDG-association and G2 arrest activities of Vpr, respectively. For ease of comparison, apoptotic activity was normalized to values obtained for the parental LAI Vpr.
Discussion
Here, we provide evidence that an unusual polymorphism identified in Vpr genes from an asymptomatic viremic patient profoundly impaired syncytium-independent viral cytopathicity and Vpr-dependent apoptosis in vitro. We postulate that such Vpr polymorphisms that uncouple viral replication and cytopathicity in vitro may have accounted, at least in part, for maintenance of CD4+ T lymphocyte homeostasis in the face of viremia in this non-progressor. Our studies further point to a direct role for Vpr in influencing lymphocyte destruction both in vitro and in vivo, implying that Vpr may harbor a determinant of viral cytopathogenicity.
Studies on individuals with non-progressive infection have provided valuable information on host factors that are important for viral replication in vivo. The presence of polymorphisms and deletions within HIV-1 second receptors CCR5 and CCR2 (reviewed in ref. 25) underscores the importance of these cellular proteins in viral replication. Similarly, polymorphisms within viral genes that appear in non-progressors underscore a role for those proteins in some aspect of viral replication or pathogenicity or both. Deletions and difficult-to-revert polymorphisms have been identified in HIV-1 Nef, Gag, Env, Rev, Vpu, and Vpr (13–15, 26–28). In all cases, non-progressors containing deletions and hard-to-revert polymorphisms in viral genes maintained low to undetectable levels of plasma viral RNA. Defects have been identified in vpr alleles obtained from a non-progressing mother and child pair (29). The influence of such defects on an activity of Vpr that could have contributed to the non-progressor phenotype was not evaluated although many of the mutations affected the tat reading frame and were likely to severely compromise viral replication capacity. Studies attempting to correlate an in vitro activity of a particular viral gene product with in vivo pathogenicity have focused on the viral envelope glycoprotein. The fusogenic activity of the envelope glycoprotein contributes both to syncytium-dependent and -independent cell killing (4, 30). Whereas destruction of CD4+ T cells in lymph nodes of SHIV-infected monkeys occurred independently of syncytium formation, viruses containing highly fusogenic envelopes were more able to effect lymphocyte depletion (31), which supports a role for envelope glycoprotein in in vivo cytopathogenicity. Earlier studies by us have similarly reported that properties of virological clones established from patient P1016 can be attributed, in part, to differences in the viral envelope glycoprotein (32). However, the in vivo prevalence of envelope polymorphisms identified in in vitro established biological clones was not determined. In the current study, a Q3R substitution at the N terminus of Vpr was sufficient to markedly impair viral cytopathic potential in the presence of a wild-type envelope glycoprotein. Therefore, we propose that, in addition to the envelope glycoprotein, Vpr qualifies as an independent cytopathogenic determinant.
Apoptosis is a host defense mechanism that restricts virus replication, and many viruses have evolved anti-apoptotic functions to preserve the longevity of the infected cell. A recent example is the anti-apoptotic property ascribed to HIV-1 Nef (33). Therefore, one might predict that polymorphisms, such as the one described here, that impair an apoptotic activity of HIV-1 would provide a replication advantage that would be selected for in the host. However, pro-apoptotic and anti-apoptotic activities have been described for a number of HIV-1 proteins, including envelope, tat, Vpr, and Nef (for review, see ref. 9). Therefore, the absolute relationship between viral apoptotic capacity and replication capacity are still unclear. High viral loads are maintained under conditions of rapid host cell turnover (34). Viruses such as HIV-1 may have evolved a rapid generation time to escape the host cell before the onset of host cell apoptosis and death. It should also be pointed out that the studies reported here do not suggest that the function of Vpr is to induce apoptosis. Rather, apoptosis may be a consequence of one of Vpr's activities as it carries out its role in promoting viral replication. It should be noted that our study refers to a single, non-progressor patient. Whereas additional studies may be required to substantiate the contribution of Vpr polymorphisms to non-progressive infection, our results point to future studies aimed at correlating rates of disease progression with cytopathic and apoptotic activities of Vpr.
Acknowledgments
We thank T. Pinkos for manuscript preparation, N. Nelson for preparation of the figures, and M. Zapp (University of Massachusetts CFAR Molecular Biology Core) for sequence analysis. The HIV-1 Vpr antibody (HIV-1NL4-3 Vpr 1–46) was provided by the National Institutes of Health AIDS Research and Reference Reagent Program (National Institute of Allergy and Infectious Diseases, Bethesda). This work was supported in part by National Institutes of Health Grants RR11589, AI32890, and AI37475 (to M. Stevenson), and AI32907 (to J.L.S.), by funding from the Edward Jenner Institute for Vaccine Research (to M. Stevenson), and by funds from the University of Massachusetts Center for AIDS Research (CFAR) to M. Somasundaran (AI42845).
Abbreviations
- SI
syncytium-inducing
- NSI
non-syncytium-inducing
- UDG
uracil DNA glycosylase
References
- 1.Mellors J W, Munoz A, Giorgi J V, Margolick J B, Tassoni C J, Gupta P, Kingsley L A, Todd J A, Saah A J, Detels R, et al. Ann Intern Med. 1997;126:946–954. doi: 10.7326/0003-4819-126-12-199706150-00003. [DOI] [PubMed] [Google Scholar]
- 2.McCune J M. Nature (London) 2001;410:974–979. doi: 10.1038/35073648. [DOI] [PubMed] [Google Scholar]
- 3.Cao J, Park I W, Cooper A, Sodroski J. J Virol. 1996;70:1340–1354. doi: 10.1128/jvi.70.3.1340-1354.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.LaBonte J A, Patel T, Hofmann W, Sodroski J. J Virol. 2000;74:10690–10698. doi: 10.1128/jvi.74.22.10690-10698.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lifson J D, Feinberg M D, Reynes G R, Rabin L, Banapour B, Chakrabarti B S, Moss B, Wong-Staal F, Steimer K S, Engleman E G. Nature (London) 1986;323:725–728. doi: 10.1038/323725a0. [DOI] [PubMed] [Google Scholar]
- 6.Bjorndal A, Deng H, Jansson M, Fiore J R, Colognesi C, Karlsson A, Albert J, Scarlatti G, Littman D R, Fenyo E M. J Virol. 1997;71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asjo B, Morfeldt-Manson L, Albert J, Biberfeld G, Karlsson A, Lidman K, Fenyo E M. Lancet. 1986;2:660–662. [PubMed] [Google Scholar]
- 8.Tersmette M, VanDongen J J M, Clapham P R, DeGoede R E Y, Wolvers-Tettero I L M, Geurts Van Kessel A, Huisman J G, Weiss R A, Miedema F. Virology. 1989;168:267–273. doi: 10.1016/0042-6822(89)90266-3. [DOI] [PubMed] [Google Scholar]
- 9.Roshal M, Zhu Y, Planelles V. Apoptosis. 2001;6:103–116. doi: 10.1023/a:1009636530839. [DOI] [PubMed] [Google Scholar]
- 10.Verhofstede C, Reniers S, Wanzeele F, Plum J. AIDS. 1994;8:1421–1427. doi: 10.1097/00002030-199410000-00008. [DOI] [PubMed] [Google Scholar]
- 11.Mellors J W. Nat Med. 1996;2:274–275. doi: 10.1038/nm0396-274. [DOI] [PubMed] [Google Scholar]
- 12.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 13.Kirchhoff F, Greenough T C, Brettler D B, Sullivan J L, Desrosiers R C. N Engl J Med. 1995;332:228–232. doi: 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]
- 14.Deacon N J, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker D J, McPhee D A, Greenway A L, Ellett A, Chatfield C, et al. Science. 1995;270:988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- 15.Alexander L, Weiskopf E, Greenough T C, Gaddis N C, Auerbach M R, Malim M H, O'Brien S J, Walker B D, Sullivan J L, Desrosiers R C. J Virol. 2000;74:4361–4376. doi: 10.1128/jvi.74.9.4361-4376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erkeller-Yuksel F M, Deneys V, Yuksel B, Hannet I, Hulstaert F, Hamilton C, Mackinnon H, Stokes L T, Munhyeshuli V, Vanlangendonck F, et al. J Pediatr. 1992;120:216–222. doi: 10.1016/s0022-3476(05)80430-5. [DOI] [PubMed] [Google Scholar]
- 17.Denny T, Yogev R, Gelman R, Skuza C, Oleske J, Chadwick E, Cheng S C, Connor E. J Am Med Assoc. 1992;267:1484–1488. [PubMed] [Google Scholar]
- 18.Swingler S, Mann A, Jacqué J-M, Brichacek B, Sasseville V G, Williams K, Lackner A A, Janoff E N, Wang R, Fisher D, Stevenson M. Nat Med. 1999;5:997–1003. doi: 10.1038/12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Selig L, Benichou B, Rogel M E, Wu L I, Vodicka M A, Sire J, Benarous R, Emerman M. J Virol. 1997;71:4842–4846. doi: 10.1128/jvi.71.6.4842-4846.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rogel M E, Wu L I, Emerman M. J Virol. 1995;69:882–888. doi: 10.1128/jvi.69.2.882-888.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirsch V M, Sharkey M E, Brown C R, Brichacek B, Goldstein S, Wakefield J, Byrum R, Elkins W R, Hahn B H, Lifson J D, Stevenson M. Nat Med. 1998;4:1401–1408. doi: 10.1038/3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakaya T, Fujinaga K, Kishi M, Oka S, Kurata T, Jones I M, Ikuta K. FEBS Lett. 1994;354:17–22. doi: 10.1016/0014-5793(94)01074-9. [DOI] [PubMed] [Google Scholar]
- 23.Stevenson M. Curr Top Microbiol Immunol. 2002;261:1–30. doi: 10.1007/978-3-642-56114-6_1. [DOI] [PubMed] [Google Scholar]
- 24.Patel C A, Mukhtar M, Harley S, Kulkosky J, Pomerantz R J. J Neurovirol. 2002;8:86–99. doi: 10.1080/13550280290049552. [DOI] [PubMed] [Google Scholar]
- 25.Michael N L. Curr Opin Immunol. 1999;11:466–474. doi: 10.1016/S0952-7915(99)80078-8. [DOI] [PubMed] [Google Scholar]
- 26.Salvi R, Garbuglia A R, Di Caro A, Pulciani S, Montella F, Benedetto A. J Virol. 1998;72:3646–3657. doi: 10.1128/jvi.72.5.3646-3657.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iversen A K, Shpaer E G, Rodrigo A G, Hirsch M S, Walker B D, Sheppard H W, Merigan T C, Mullins J I. J Virol. 1995;69:5743–5753. doi: 10.1128/jvi.69.9.5743-5753.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Binley J M, Jin X, Huang Y, Zhang L, Cao Y, Ho D D, Moore J P. J Virol. 1998;72:3472–3474. doi: 10.1128/jvi.72.4.3472-3474.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang B, Ge Y C, Palasanthiran P, Xiang S-H, Ziegler J, Dwyer D E, Randle C, Dowton D, Cunningham A, Saksena N K. Virology. 1996;223:224–232. doi: 10.1006/viro.1996.0471. [DOI] [PubMed] [Google Scholar]
- 30.Cao Y, Qin L, Zhang L, Safrit J, Ho D D. Immunol Lett. 1996;51:7–13. doi: 10.1016/0165-2478(96)02548-5. [DOI] [PubMed] [Google Scholar]
- 31.Etemad-Moghadam B, Rhone D, Steenbeke T, Sun Y, Manola J, Gelman R, Fanton J W, Racz P, Tenner-Racz K, Axthelm M K, et al. J Virol. 2001;75:5646–5655. doi: 10.1128/JVI.75.12.5646-5655.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forte S E, Somasundaran M, Sullivan J L. AIDS Res Hum Retroviruses. 2000;16:125–137. doi: 10.1089/088922200309476. [DOI] [PubMed] [Google Scholar]
- 33.Geleziunas R, Xu W, Takeda K, Ichijo H, Greene W C. Nature (London) 2001;410:834–838. doi: 10.1038/35071111. [DOI] [PubMed] [Google Scholar]
- 34.Mittler J E, Markowitz M, Ho D D, Perelson A S. AIDS. 1999;13:1415–1417. doi: 10.1097/00002030-199907300-00023. [DOI] [PubMed] [Google Scholar]