Abstract
Pancreatic insufficiency (PI) when left untreated results in a state of malnutrition due to an inability to absorb nutrients. Frequently, PI is diagnosed as part of a larger clinical presentation in cystic fibrosis or Shwachman–Diamond syndrome. In this study, a mouse model for isolated exocrine PI was identified in a mouse line generated by a transgene insertion. The trait is inherited in an autosomal recessive pattern, and homozygous animals are growth retarded, have abnormal immunity, and have reduced life span. Mice with the disease locus, named pequeño (pq), exhibit progressive apoptosis of pancreatic acinar cells with severe exocrine acinar cell loss by 8 wk of age, while the islets and ductal tissue persist. The mutation in pq/pq mice results from a random transgene insertion. Molecular characterization of the transgene insertion site by fluorescent in situ hybridization and genomic deletion mapping identified an approximately 210-kb deletion on Chromosome 3, deleting two genes. One of these genes, Serpini2, encodes a protein that is a member of the serpin family of protease inhibitors. Reintroduction of only the Serpini2 gene by bacterial artificial chromosome transgenic complementation corrected the acinar cell defect as well as body weight and immune phenotypes, showing that deletion of Serpini2 causes the pequeño phenotype. Dietary supplementation of pancreatic enzymes also corrected body size, body weight, and immunodeficiency, and increased the life span of Serpini2-deficient mice, despite continued acinar cell loss. To our knowledge, this study describes the first characterized genetic animal model for isolated PI. Genetic complementation of the transgene insertion mutant demonstrates that Serpini2 deficiency directly results in the acinar cell apoptosis, malabsorption, and malnutrition observed in pq/pq mice. The rescue of growth retardation, immunodeficiency, and mortality by either Serpini2 bacterial artificial chromosome transgenic expression or by pancreatic enzyme supplementation demonstrates that these phenotypes are secondary to malnutrition in pq/pq mice.
Synopsis
Pancreatic insufficiency is defined by the inability to digest and absorb nutrients due to the loss of pancreatic enzyme function or loss of the acinar cells that produce the enzymes. In this manuscript the authors have described a mouse model of pancreatic insufficiency characterized by the specific loss of pancreatic acinar cells. This specific acinar cell loss results in mice that are unable to digest and absorb nutrients from the diet, stunting the animal's growth and giving rise to immunological anomalies. The authors have identified a serendipitous transgene insertion/deletion encompassing the mouse Serpini2 gene locus as the source of the phenotypes observed. Reintroduction of the Serpini2 gene, a member of the serpin family of serine cysteine protease inhibitors, by bacterial artificial chromosome complementation corrects the pancreatic and immunological phenotypes of the disorder, confirming Serpini2 as the responsible gene. Reintroduction of pancreatic enzymes through diet supplementation is also capable of correcting the reduction in size and weight, reduction in viability, and immunological deficiencies, indicating that these phenotypes are secondary to malnutrition alone. This work provides a new mouse model for investigation of malnutrition/malabsorption due to pancreatic insufficiency and identifies a novel function for the serpin family member Serpini2.
Introduction
The ability to ectopically express novel genes in the mouse genome via introduction of transgenic sequences has proved to be a valuable tool in the study of mammalian development and disease. However, in approximately 5% of the transgenic lines produced, an insertional mutation at the genomic integration locus also occurs [1]. Often the insertion of the transgene is accompanied by a loss of surrounding genomic sequences and, dependent on the location of nearby coding and regulatory sequences, will produce an observable phenotype. We have been able to take advantage of such a transgene insertion/deletion event to identify and characterize a mouse model of pancreatic insufficiency (PI).
PI or a lack of pancreatic enzymes can lead to malabsorption due to an inability to digest and absorb nutrients. The most common genetic disorder that exhibits PI is cystic fibrosis (CF), as 95% of patients with CF have pancreatic acinar cell loss, with fatty cell replacement and interstitial fibrosis [2]. A second example of a syndrome that involves PI is the Shwachman–Diamond syndrome (SDS). Patients present with PI due to pancreatic acinar cell loss, but also have clinical manifestations of short stature, neutropenia, pancytopenia and predisposition to acute myelogenous leukemia [3].
Animal models of acute PI have been created by inducing pancreatic acinar cell apoptosis, either by administration of the synthetic cholecystokinin analog caerulein [4] or by physical blockage of the pancreas by ductal ligation [5]. However, defined genetic animal models of PI have been lacking. Only recently has a genetic model for an inbred CF mouse been described that recapitulates pancreas involvement of the disorder; however, this animal also manifests other features of CF [6].
In this paper we describe a new mouse model for exocrine PI that arose serendipitously in a line of mice carrying a transgene insertion. This insertion event resulted in a genomic deletion of the gene encoding the protease inhibitor SERPINI2/ PANCPIN. The mutant allele, termed pequeño (which is the Spanish word for “small”), is characterized by severe exocrine acinar cell loss at 8 wk of age, while islets and ductal tissue are spared. The disorder is inherited in an autosomal recessive pattern, and untreated homozygotes are malnourished, with a body weight one-third smaller than control littermates. Secondary to the malnutrition, these animals also have compromised immunity and a reduced life span. Administration of pancreatic enzyme diet supplementation is sufficient to reverse the effects of malnutrition in pq/pq animals, counteracting the growth defects, decreased viability, and immunodeficiency.
Results
Identification and Pathology of the Pequeño Mouse Line
In offspring of a p3pTVA-B line of transgenic mice that were bred to homozygosity, we observed runted mice with a reduced viability phenotype that we called pequeño (Figure 1A). Line p3pTVA-B was one of five independent transgenic mouse lines generated using the Pax3 promoter to drive expression of the avian retroviral receptor tv-a [7]. The four other lines generated did not produce the pequeño phenotype, suggesting that it was the site of transgene insertion that caused the phenotype. Analyses of 34 progeny resulting from the intercross of two obligate heterozygous mice demonstrated that 24 transgenic offspring (71%) were produced, in line with the expected Mendelian ratio. Six of the transgene-positive mice (6/34, 17.6%) were significantly smaller at weaning, suggesting that they were homozygous for a recessive mutation. Fluorescent in situ hybridization (FISH) analyses of metaphase spreads from three pequeño animals using the p3pTVA construct as a probe were performed. All three mice were homozygous for the transgene insertion on mouse Chromosome 3 (data not shown). We concluded that the pequeño phenotype is due to a homozygous mutation (pq/pq) that is linked to the transgene insertion and that the phenotype is inherited in an autosomal recessive pattern, as the heterozygotes had no apparent manifestations.
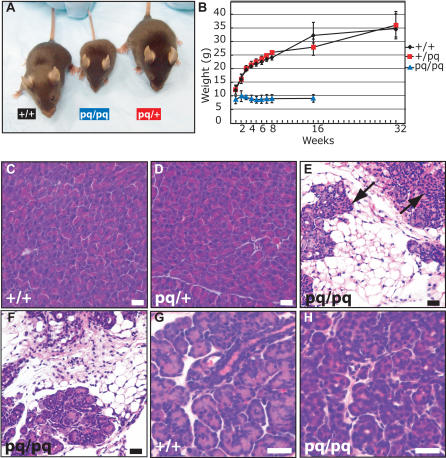
Figure 1. Acinar Cell Loss in pequeño Mice.
(A) Size of pq/pq mice in relation to wild-type and heterozygous littermates at 8 wk of age.
(B) Average weight of n = 6 males for each genotype over a 32-wk period.
(C–H) Pancreas hematoxylin and eosin histological analyses were performed on 8-wk-old (C, D, and E), 3-wk-old (F), and 1-wk-old (G and H) mice. Wild-type (+,+) acinar cells (C) and heterozygous (pq/+) acinar cells (D) at 8 wk of age were morphologically normal. This is in contrast to homozygous (pq/pq) animals at 8 wk of age (E) and 3 wk of age (F), where there is a severe loss of exocrine pancreatic acinar cells while duct cells and islet cells (arrow) are normal. Acinar cells are present in pancreas samples from 1-wk-old pq/pq mice (H), but the cells are smaller and show reduced cytosolic volume when compared to wild-type littermates (G). Bar represents 25 μm.
The pq/pq mice were on average one-third the weight of wild-type littermates (Figure 1B) and produced a gray-colored stool. Pathological assessment at 8 wk of age revealed that the spleen and thymus were dramatically reduced in size and the pancreas was abnormal by histological analyses of affected animals when compared to unaffected littermates. Gross skeletal abnormalities were not visible by X-ray or skeletal preparation (data not shown). The thymus of homozygous mutants (n = 6) were about 4-fold smaller than wild-type littermates (n = 6) (2.90% versus 11.44% thymus/brain weight; p < 2.5 × 10−5). Similar results were observed for the spleen (4.05% versus 16.7% spleen/brain weight; p < 9.7 × 10−7). Histological examination of the endocrine pancreas for pq/pq mice revealed that islets and ductal tissue appeared normal. Analysis of the exocrine pancreas showed a striking loss of acinar cells (Figure 1). Histological evaluation of the pancreas of younger animals at 1 wk of age showed abnormal acinar cells that contained fewer zymogen granules (ZGs) than those of littermate controls (Figure 1G and 1H). However, by 3 wk of age pq/pq animals had only about one-half the number of the pancreatic acinar cells when compared to wild-type, despite the normal appearance of ductal and endocrine islet cells (Figure 1F). This observation indicated that extensive acinar cell loss was occurring between 1 and 3 wk of age in pq/pq animals.
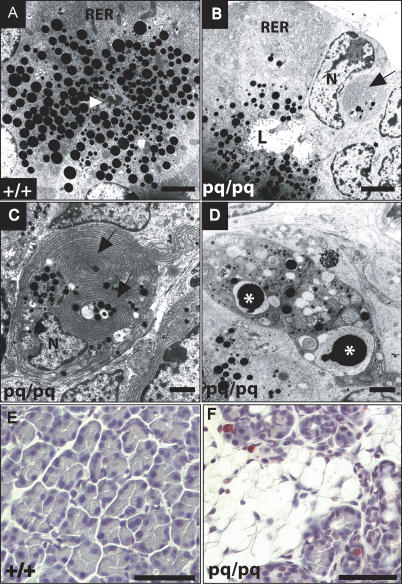
Further analyses of the pancreas from 1-wk-old pq/pq animals by electron microscopy revealed a reduction in size and number of dense ZGs in the cytoplasm of the pq/pq acinar cells and a lack of dense material within the ductal network (Figure 2). In contrast, the pancreas of wild-type littermates contained many ZGs, and electron-dense material representing large quantities of enzymes is found in the pancreatic ducts (Figure 2A). Additionally, many pq/pq acinar cells were found to exhibit hallmark features of cells undergoing apoptosis. These acinar cells were smaller than adjacent cells and contained compact cytoplasmic organelles, condensed nuclear chromatin, and concentrically whirled bodies of rough endoplasmic reticulum (Figure 2B–2D). In addition, these acinar cells were observed being phagocytosed by neighboring epithelial cells while some were sloughed directly into the ductal lumina (Figure 2B–2D) [8]. As a second independent method of assessing apoptosis, pancreatic tissue was analyzed for DNA fragmentation using terminal deoxynucleotidyl transferase biotin–dUTP nick end labeling (TUNEL) staining. Pancreas sections from two animals in each group (wild-type and pq/pq) were counted for TUNEL staining in 12 independent, randomly selected fields (Figure 2E and 2F). The number of TUNEL-positive cells for pq/pq animals (mean counts of 7.2 and 4.9 cells per field) was significantly increased when compared to that of the wild-type (0.33 and 0.25 cells per field). Analyses of the four pair-wise comparisons were statistically significant (p < 0.01). Taken together, these results are consistent with apoptosis as the major cause of acinar cell loss.
Figure 2. Electron Microscopy Shows pequeño Acinar Cells Are Present but Undergoing Apoptosis at 1 Wk of Age.
(A) Wild-type acinar cells have normal cellular morphology and contained abundant numbers of ZGs (seen as black circular vesicles containing electron-dense material), and electron-dense material from ZGs is present in the ductal lumen (white arrow).
(B–D) pq/pq mice have fewer and smaller ZGs and no dense material in the ductal lumen (L) when compared to wild-type animals. A significant number of acinar cells are undergoing apoptosis, as indicated by the presence of cellular phagocytosis (C and D), rough endoplasmic reticulum whirling bodies (B and C) (black arrows), and nuclear condensation and fragmentation (D) (asterisk).
(E and F) TUNEL staining of 3-wk-old pancreas tissue from wild-type (E) and pq/pq (F) mice indicates an increase in TUNEL-positive, red-fushcin-staining cells in pq/pq mice as compared to wild-type.
L, lumen; N, nucleus; RER, rough endoplasmic reticulum. For (A–D) bar represents 5 μm. For (E and F) bar represents 50 μm.
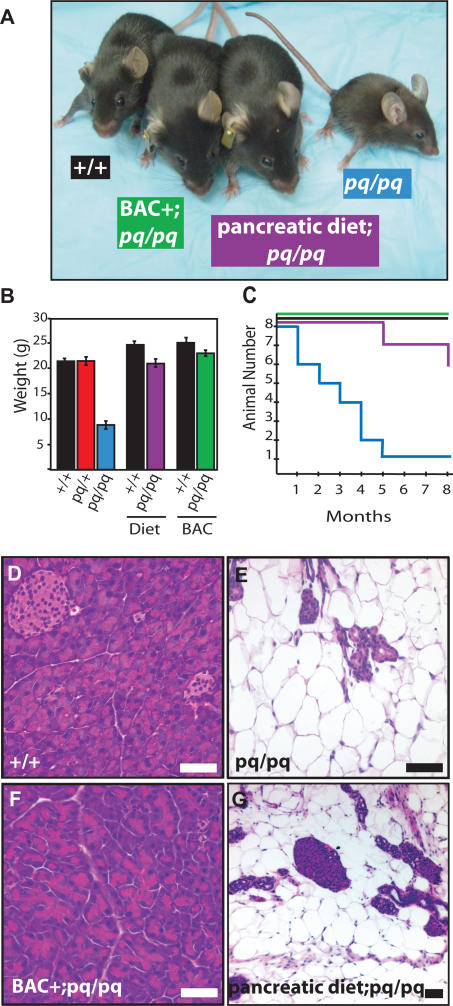
To test the hypothesis that growth failure is caused by PI, pq/pq animals were treated with a dietary supplementation of pancreatic enzymes in addition to a normal diet and assessed for correction of the malnutrition (Figure 3). Animals supplemented with a pancreatic enzyme diet were monitored for size, weight, and viability. Treated pq/pq mice gained both weight and length when compared with untreated pq/pq animals (Figure 3A and 3B). As expected, the enzyme supplement did not correct the acinar cell deficiency (Figure 3G). We conclude that the PI alone was responsible for the growth retardation in pq/pq mice (Figure 3).
Figure 3. pequeño Malnutrition Is Rescued by BAC Transgenic Expressing Serpini2 Gene or Pancreatic Enzyme Diet Supplementation.
(A–C) Treatment of pq/pq mice (blue) pharmacologically with daily pancreatic enzyme diet supplement (purple) or genetically with BAC 150E1 transgene (green), rescues (A) size, (B) weight, and (C) animal viability in comparison to wild-type mice (black). For (B) weight measurements are averaged from n = 6 males from each genotype.
(D–G) Hematoxylin and eosin staining of pancreas sections from 8-wk-old mice. Wild-type mice (D) have normal pancreatic acinar cells present and normal pancreatic morphology while in (E) pq/pq mice there is a loss of acinar cells. In the BAC+; pq/pq mice (F), pancreas acinar cells are present. The acinar cell loss in pq/pq mice treated with enzyme supplementation (G) is the same as untreated pq/pq mice (A–C), although diet supplementation is effective in combating the effects of malnutrition. Bar represents 25 μm.
Identification of the Gene Responsible for the Pequeño Phenotype
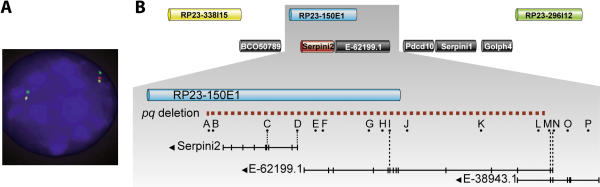
To refine the location of the transgene insertion site in pequeño mice, the p3pTVA transgene was hybridized to heterozygous pq/+ chromosome spreads in combination with bacterial artificial chromosomes (BACs) spanning Chromosome 3 using multicolor FISH. BACs spanning 52,290,000–85,000,000 bp of Chromosome 3 were identified with NCBI Map Viewer (http://www.ncbi.nlm.nih.gov/mapview/) mouse build 31, and obtained from Children's Hospital Oakland Research Institute (http://www.chori.org/bacpac). This analysis showed that the BAC clones RP23-388I15 and RP23-296I12 flanked the p3pTVA transgene insertion and mapped the insertion to a 2.3-MB region (Figure 4). This region contained four genes of known function and several putative genes represented by expressed sequence tags and/or gene prediction algorithms (Figure 4B). One candidate gene in the region, Serpini2, a serpin protein family member, is predicted to be a serine-cysteine protease inhibitor. In addition, Serpini2 has been shown to be expressed at high levels in pancreas and adipose tissue [9,10], making it an excellent candidate gene for the pequeño phenotype.
Figure 4. Pequeño Phenotype Is the Result of a 210-kb Genomic Deletion.
(A) FISH to interphase pq/+ chromosome spreads with a p3pTVA transgene probe (red) locates the transgene insertion on mouse Chromosome 3 between BACs RP23-338I15 (yellow) and RP23-296I12 (green).
(B) The genomic region defined by BAC FISH localization contained six known or predicted genes, one of which, Serpini2, is highly expressed in pancreas and adipose tissue. PCR primer sets A–P (Table S1) were used to characterize and define the identified ~210-kb deletion, indicated by red dashed line. PCR primer sets that correspond to known and predicted exon sequences are indicated by black vertical dashed lines. The genomic deletion encompassed the entire Serpini2 gene locus and exons 4 to 17 of the predicted gene E-62199.1/E-38943.1.
The genomic draft sequence from the mouse February 2003 freeze build at the University of California at Santa Cruz (http://genome.ucsc.edu/cgi-bin/hgGateway) was used to design primers within the Serpini2 gene and to the flanking genomic DNA (Figure 4B; Table S1). PCR analysis established that the transgene insertion caused in an approximately 210-kb genomic deletion at the locus, which included the entire Serpini2 gene and continued distally to include several 3′ exons of Ensembl-predicted novel gene transcript sequences ENSMUST00000062199.1 (E-62199.1) and ENSMUST00000038943.1 (E-38943.1) (Figure 4B). The size and location of the deletion established the possibility that the pequeño phenotype could be a recessively inherited contiguous gene syndrome.
We reasoned that the restricted expression of the Serpini2 gene in pancreas and adipose tissue as well as its proposed function as a serine protease inhibitor suggested that deletion of the Serpini2 locus alone caused the pequeño phenotype. An alternative explanation was that deletion of the two Ensemble-predicted transcript sequences E-62199.1 and E-38943.1 contributed to the phenotype. Sequence analysis of the predicted exons for E-62199.1/E-38943.1 suggested they are a single gene and an ortholog of the human locus Unigene cluster Hs.376728. The human gene sequence and predicted mouse gene sequences are identical in exon–intron structure and are 75% identical at the amino acid level. The pequeño deletion includes the entire Serpini2 gene locus and exons 14 to 17 of E-62199.1/E-38943.1, leaving the first three exons intact.
BAC Transgene Complementation Cross Rescues Pequeño Acinar Cell Defect
In order to dissect the role of Serpini2 and rule out the predicted genes as candidates for pequeño, we produced a BAC transgenic line, 150E1, to perform Serpini2 complementation crosses with pequeño mice. BAC RP23-150E1 was selected using the University of California at Santa Cruz February 2003 browser because it contained the entire intact Serpini2 gene and 71 kb of the upstream promoter region, but not a functional E-62199.1/E-38943.1 gene. BAC-positive (BAC+) transgenic animals were crossed with pq/+ mice and the resulting double heterozygous BAC+; pq/+ animals were backcrossed to pq/+ animals. Animals were genotyped with primers to the p3pTVA transgene, a deletion-specific primer “77kb” (within the genomic deletion, but outside of BAC 150E1), and BAC vector end-specific primers. Transgenic BAC+; pq/pq animals were indistinguishable from BAC−; +/+ and BAC+; +/+ animals by size, weight and viability (see Figure 3A–3C). RT-PCR from pancreas RNA obtained from wild-type, BAC−; pq/pq null, and BAC+; pq/pq BAC-complemented animals was performed. The results demonstrated that Serpini2 is not expressed in BAC−; pq/pq null animals and that the BAC transgene in BAC+; pq/pq animals restored expression of Serpini2 in pancreas (data not shown). Histological analysis of BAC+; pq/pq animals revealed they had normal acinar cells, showing that Serpini2 expression alone corrected the acinar cell defect phenotype (Figure 3D–3F). We conclude that deletion of the Serpini2 gene, and not E-62199.1/E-38943.1, causes the pequeño phenotype of acinar cell loss leading to PI and subsequent growth retardation due to malabsorption and malnutrition.
Immune Cell Defects in pq/pq Mice
The pq/pq mice without pancreatic diet supplementation exhibited decreased life span (typically 2–7 mo compared to over 1 y for normal mice). Interestingly, when pq/pq mice without pancreatic diet supplementation were housed in sterile cages with acidified water, their condition improved and their life span increased compared to pq/pq animals housed in standard conditions. This suggested that the pq/pq mice had compromised immune function and infections were contributing to the morbidity (data not shown). We therefore sought to characterize the immune system of the pq/pq mice.
Analyses of lymphoid organs revealed that pq/pq mice had a reduction in thymic, splenic, and bone marrow cellularity by 21 and 28 d after birth (Figure 5). In particular, spleens from pq/pq mice at 3–4 wk of age showed a dramatic reduction in the number of lymphocytes present, with T cell numbers reduced approximately 5-fold and B cell numbers reduced more than 10-fold. Thymocyte numbers were also severely reduced (10- to 20-fold), suggesting that the decrease in mature T cells resulted from reduced thymic output. Despite the dramatic reduction in thymic size, pq/pq mice had a normal thymocyte profile, with cells progressing from the most immature double negative (CD4−8−) population to double positive CD4+8+ cells and both CD4 and CD8 single-positive mature cells, with no clear block at any stage (Figure 6). In particular, the increase in the percentage of single-positive CD4+ and CD8+ cells argues that the cells exhibit no defects in selection and can progress to the most mature stages of thymic development. These results suggested that thymic development was not blocked per se, but that there was reduced cell input or viability at multiple stages.
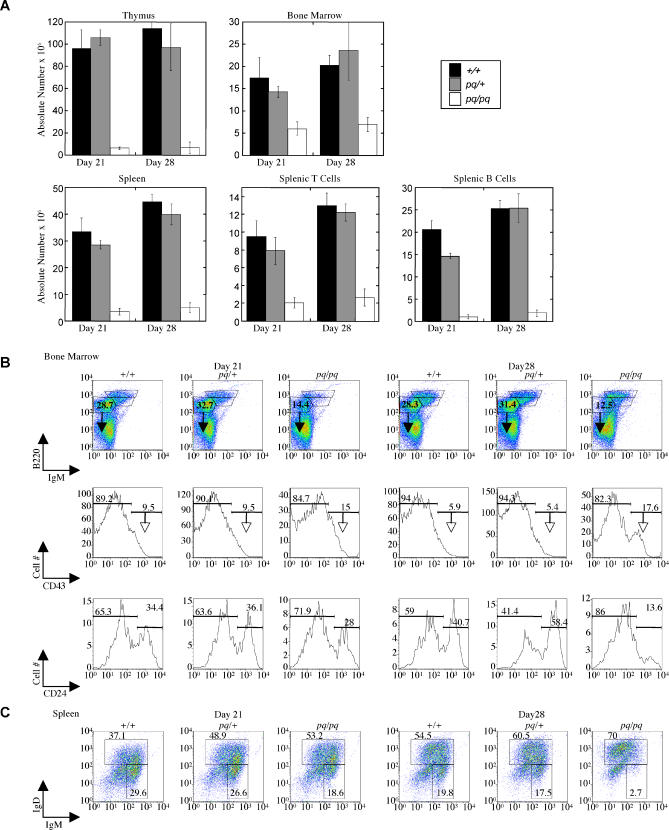
Figure 5. Decreased Lymphocyte Numbers and Impaired B cell Development in pequeño Mice.
(A) pequeño mice exhibit a striking reduction in the cellularity of the thymus, spleen and bone marrow at days 21 and 28 of life (n = 3).
(B) pequeño mice show decreased early B cell progenitors in the bone marrow. Expression profiles of B220 and IgM on total lymphoid cells in bone marrow are shown for wild-type (top row, percentage of B220+IgM− pro-/pre-B cell fraction are indicated). The B220+IgM− pro-/pre-B cell fraction (filled arrow) was gated to analyze the developmental stages using CD43 (middle row). The B220+IgM−CD43+ (less mature) fraction (open arrow) was then gated on to analyze CD24 expression (bottom row). pq/pq mice exhibit an increase in the proportion of CD43+CD24− B cells, indicating a block in the pre-/pro-B to early pro-B transition.
(C) Flow cytometric analysis of surface IgM+/IgD− expression on B220+ cells in the spleen indicates a decreased percentage of immature (IgM+/IgD−) B cells in the pequeño mice.
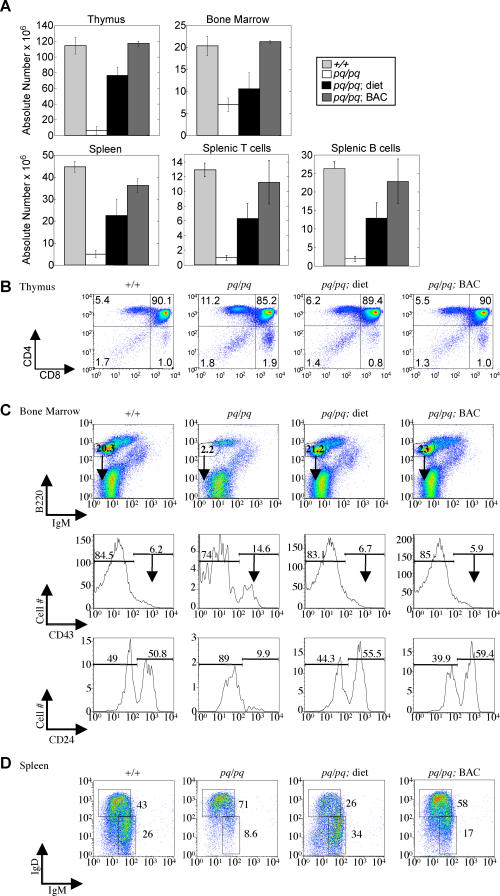
Figure 6. Pancreatic Enzyme Supplementation and BAC Transgenesis Rescue Lymphocyte Cells Numbers and Developmental Defects in pequeño Mice.
(A) pequeño mice with either enzyme supplementation (pq/pq; diet) or a BAC transgenic (pq/pq; BAC) display increased cellularity of the thymus, spleen, and bone marrow at day 28 of life (n = 4).
(B–D) Pancreatic enzyme supplementation and the BAC transgenic rescue the developmental defects in the pequeño mice. (B) CD4 and CD8 expression profiles of the thymus. (C) Expression profiles of B220 and IgM on total lymphoid cells in bone marrow (top row). The B220+IgM− pro-/pre-B cell fraction (filled arrow) was gated to analyze the developmental stages using CD43 (middle row). The B220+IgM−CD43+ (less mature) fraction (open arrow) was then gated on to analyze CD24 expression (bottom row). (D) Splenic expression profile analysis of surface IgM/IgD expression on B220+ cells .
We also observed a defect in B cell development. B lymphocytes develop in the bone marrow through distinct stages characterized by differential expression of cell surface markers and the ordered rearrangement of Ig heavy and light chain gene segments [11,12]. Wild-type mice have populations of B220+sIgM− pro- and pre-B cells in their bone marrow that upregulate and express increasing levels of sIgM on their cell surfaces as they develop into immature and mature B cells. In contrast, pq/pq mice had fewer pro- and pre-B cells (B220+IgM−), and expression of IgM on immature cells was lower, particularly at day 28 (see Figure 5B). Within the B220+sIgM− fractions, the most immature pro-B cell fractions, fractions A–C, are further defined by expression of CD43 (B220+IgM−CD43+) and differing expression of CD24 [13]. Fraction A is CD24− and contains cells that first appear to be committed to the B cell lineage (pre-pro-B cells). Fractions B (early pro-B cells) and C (late pro-B cells) now express CD24 and initiate rearrangement at the IgH locus (B220+IgM−CD43+CD24+). Compared to wild-type, pq/pq pre-B cells manifested a higher percentage of B220+IgM−CD43+ cells, indicative of a partial block in B cell development prior to the pre-B cell stage. This block was more apparent as the mice aged (compare day 28 to day 21, Figure 5B). Moreover, within the CD43+ fraction, pq/pq pro-B cells expressed lower levels of CD24, demonstrating that this block occurred between the pre-proB and early pro-B cell stage. Similarly, in the spleen, reduced numbers of the most immature population of B cells (IgM+IgD−) were observed (Figure 5C), arguing that there was reduced production of B cells in the bone marrow.
Pancreatic Diet Supplementation and BAC Transgene Rescues Immune System Defects
To determine whether the PI and subsequent malnourished state alone were contributing to the impaired immune system or whether there was an intrinsic defect in pq/pq lymphocytes due to the lack of SERPINI2, we sought to determine how the pq/pq mice would respond to dietetic supplementation with pancreatic enzymes in contrast to BAC+; pq/pq and pq/pq untreated animals. The pancreatic enzyme replacement therapy restored immune cell numbers in the pq/pq mice, suggesting immune system defects were secondary to malnutrition. Similarly, BAC+; pq/pq transgenic mice were also able to complement immune system defects. At 4 wk of age, enzyme supplementation resulted in a partial rescue, while animals with the BAC transgene were indistinguishable from wild-type animals with respect to cellularity of the spleen, thymus, and bone marrow and the numbers of mature T and B cells within the spleen (see Figure 6A). Moreover, both the diet supplementation and the BAC transgene insertion complemented bone marrow differentiation defects. pq/pq mice with either pancreatic enzyme diet supplementation or the BAC transgene had an increased B220+IgM− fraction and normalized development of IgM+ immature B cells and the percentage of early progenitor B cells, as defined by CD43 and CD24, when compared to pq/pq animals without either enzyme supplementation or BAC transgene introduction (Figure 6C). Splenic B cell profiles were also normalized, with increased percentages of early newly emigrated B cells (Figure 6D) in enzyme-treated and BAC transgenic pq/pq animals. Thus, the immune defects in pq/pq mice appear to be the in large part secondary to pancreatic exocrine deficiency, which leads to malnourishment caused by malabsorption.
Discussion
We have identified a mutant with PI caused by a deletion of the protease inhibitor Serpini2. This phenotype (pequeño) is inherited in an autosomal recessive pattern and is to our knowledge the first published mouse mutant to genetically model PI. The mutation, caused by a transgene insertion, led to an approximately 210-kb genomic deletion that completely removes the Serpini2 gene and a portion of the predicted gene E-62199.1/E-38943.1. Reintroduction of the Serpini2 gene alone by BAC complementation corrected the acinar cell defect and improved immune function, demonstrating that deletion of the Serpini2 gene is the primary genetic cause of the pequeño mouse phenotype.
Serpini2 was initially identified and implicated as a potential tumor suppressor gene because it was downregulated in pancreatic cancer cell lines when compared to normal pancreas [9]. Serpini2 is also known as MEPI, a gene expressed in normal breast myoepithelial cells, but not in malignant breast carcinoma cells [14]. We did not observe any overt phenotype in the pequeño mice with regard to mammary gland development/function. Female pq/pq mice that had been maintained on a pancreatic enzyme diet supplementation were able to rear litters of 8–10 pups with no difficulty. It is possible that loss of expression in mammary tissue does not affect mice mammary gland function. Alternatively, there is the potential that another serpin family member may be compensating for the loss of Serpini2 activity, as has previously been observed for loss of Serpinb6 function resulting in upregulation of Serpinb1 expression [15].
SERPINI2 is a member of the serpin superfamily of proteins that have been implicated in a variety of functions including blood coagulation, angiogenesis, inflammation, and programmed cell death [16]. In humans, 33 serpins have been identified [17]. These proteins are further subdivided into two classes, inhibitory and non-inhibitory. The former group, which includes SERPINI2, is characterized by conformational structure changes that allow for formation of covalent acyl-enzyme intermediates with the proteins' target protease, and thus have been termed “suicide substrates” [18]. The target substrates for SERPINI2 are unknown.
Further insight into the cellular location and function of SERPINI2 has come from work with the rat homolog, ZG-46p. SERPINI2/ZG-46p has been characterized in the pancreas, located predominately in the Golgi and in ZGs [19], and was isolated from ZG membranes [20]. ZGs in the acinar cells contain inactive proenzyme precursors for digestive enzymes including trypsin, amylase, chymotrypsin, and carboxypeptidase. Normal acinar cells produce large quantities of granules that are transported from the trans-Golgi network to the luminal side of the acinar cell and are then secreted to the ductal lumen upon hormonal and neural signaling mechanisms. The contents of the ZGs are transported via ducts to the duodenum (pp. 429–430 of [21]), where these enzymes are required for proper digestion and absorption of food.
Given the subcellular location of SERPINI2 in the Golgi and ZGs, in addition to the predicted gene function as a serine protease inhibitor, we hypothesize that SERPINI2 may play a role in regulating the level of inactive zymogen precursors. The importance of tight regulation of active protease levels by protease inhibitors in the exocrine pancreas is well documented. This regulation is demonstrated by considering mutations in two genes that are mutated in human hereditary pancreatitis: the cationic trypsinogen gene PRSS1 and serine protease inhibitor Kazal type 1 (SPINK1). For a small number of individuals with chronic PI, mutations in either gene have been shown to alter this balance and lead to autodigestion of tissue with inflammation [22,23]. However, genetic animal models for these two genes are lacking.
It is also important to note that differences in the severity of pancreatitis may be regulated by the type of cellular response to the inflammation, whether apoptosis or necrosis. In several models of PI, acinar cell apoptosis causes a less severe pancreatitis phenotype [24–26], in contrast to the more severe pancreatitis that has been documented with acinar cell necrosis [25,27]. It is interesting to speculate that other less severe alleles than the null allele present in the pequeño mouse model may not have severe apoptosis and may demonstrate a pancreatitis phenotype similar to that observed in patients with SPINK1 mutations.
Alternatively, SERPINI2 may function to allow proper sorting and/or transport of ZGs from the Golgi to the ductal lumen of the acinar cell. This hypothesis is consistent with the observations made of the acinar cells of 1-wk-old pq/pq mice that have yet to undergo apoptosis. In these acinar cells there is a dramatic loss of ZGs present in the cell and the ZGs appear smaller in size than those in wild-type and heterozygous littermates. Further analysis is required to differentiate these potential modes of action for SERPINI2.
It is clear that malnutrition is the primary cause of the observed pequeño phenotypes. The inability of pq/pq mice to adequately digest and absorb food from a young age adversely affects growth and immune system function. A large body of evidence argues that malnutrition, stress-induced hormones, and corticosteroids can adversely affect immune cell numbers and survival [28]. In addition, leptin, a pleiotropic molecule that regulates food intake and metabolic endocrine functions, also plays a role in immune and inflammatory responses [29]. Although the defects we observed here do not completely correlate with those associated with increased steroid exposure in mice, most studies of steroid exposure have examined short-term glucocorticoid exposure [30]. However, over-expression of a glucocorticoid receptor in the thymus produced results similar to ours, with reduced cellularity observed at multiple stages of T cell development [31]. Similar long-term studies have not examined B cell numbers or function. While we cannot rule out a direct effect of SERPINI2 on lymphocytes, the improvement of immune function by pancreatic diet supplementation strongly suggests that these defects are secondary to malnutrition, which could result in increased stress hormone and steroid production, decreased leptin levels, and/or a lack of nutrients.
It is interesting that growth retardation, PI, and immunodeficiency are present in both the pequeño mouse and the human disorder SDS. The human disease is a rare and clinically heterogeneous disorder that manifests PI, short stature, lipidosis of the pancreas, neutropenia, pancytopenia, and predisposition to acute myelogenous leukemia. Although we have not observed neutropenia in the blood of pq/pq mice (Figure S1; Table S2), it may be difficult to compare secondary immune defects in mouse and human, as effects of malnutrition and stress hormones may differ between the species. Nonetheless, the combination of PI and immune defects in this mouse mutant is intriguing, given that several recent studies indicate SDS may exhibit genetic heterogeneity [32–34].
The most common mutation for SDS has been identified and is a gene conversion caused by recombination of the ubiquitously expressed SBDS gene with the pseudogene SBDSP located 5.8 Mb away on Chromosome 7 [32]. The SBDS protein is predicted to be an RNA-processing enzyme based upon sequence homology to a yeast ortholog and cluster analysis data from microarray experiments [32]. Given the overlap in phenotypes of the pequeño mice and genetic heterogeneity predicted for patients with SDS, Serpini2 is a candidate gene for analysis in the subset of patient samples in which no mutation in the SBDS gene can be identified.
Materials and Methods
Pancreatic enzyme diet.
The pancrezyme diet was provided as a supplementation to normal mouse chow Rodent NIH-31 Auto18–4 (Ziegler, Gardeners, Pennsylvania, United States). The supplement (Pancrezyme, Daniels Pharmaceuticals, St. Petersburg, Florida, United States) was mixed with Transgenic Dough diet (Bio-Serv, Frenchtown, New Jersey, United States) at a ratio of 0.9 g supplement/50 g of transgenic dough. Five grams per animal of supplemented transgenic dough mixture was provided once every 24 h.
FISH.
Metaphase spreads were performed with standard air-drying technique from blood collected from retro-orbital bleeds and mouse spleens [35]. DNA was labeled by nick translation technique, essentially as described by Pinkel et al. [36] and Lichter et al. [37] with Spectrum Orange (Vysis, Downers Grove, Illinois, United States). On each slide 200 ng of labeled probe was applied. Repeat sequences were blocked with 10× excess Cot1 DNA, 10 μl of a hybridization mixture containing the labeled DNA, 50% formamide, 2× SSC, and 10% dextran sulfate (pH 7) and were denatured at 75 °C for 10 min, then incubated at 37 °C for 30 min. Slides were incubated for at least 1 h at 37 °C in 2× SSC, then dehydrated with 2-min ethanol washes (70%, 80%, and 90%). Slide denaturation was performed in 70% formamide/2× SSC for 2 min, then dehydrated at −20 °C in 2-min ethanol washes (70%, 80%, 90%, and 100%). Post-hybridization washes were performed at 45 °C as follows: (1) 50% formamide and 2× SSC for 15 min, (2) 0.1× SSC for 10 min. Slides were counterstained with DAPI-Antifade (250 ng/ul BM, Vector, Burlingame, California, United States).
Production of BAC transgenic mice.
P3pTVA-transgenic mice were generated as described [7]. For BAC complementation analysis, fertilized oocytes obtained from matings of C57BL/6J mice were used for pronuclear injection of RP23-150E1 BAC DNA. BAC DNA was purified by CsCl gradient isolation and not linearized prior to injection. The concentrations of BAC DNA used for microinjection were 0.5–0.75 μg/ul. A total of 69 pups were born, five of which were positive for the BAC by PCR.
Histology and electron microscopy.
Tissues evaluated by hematoxylin and eosin staining and light microscopy were fixed in 10% neutral buffered formalin routinely processed, embedded into paraffin, sectioned at 5 μm thick, and stained with hematoxylin and eosin, performed by Histoserve (Rockville, Maryland, United States). Pancreas tissue evaluated by electron microscopy was fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4) overnight. The tissue was washed with cacodylate buffer and postfixed with 1% OsO4 for 2 h. Tissue was washed again with 0.1 M cacodylate buffer, serially dehydrated in ethanol and propylene oxide and embedded in Eponate 12 resin (Ted Pella, Redding, California, United States). Thin sections, approximately 80 nm, were obtained by utilizing the Leica Ultracut-UCT ultramicrotome (Leica, Deerfield, Illinois, United States) and placed onto 400 mesh copper grids and stained with saturated uranyl acetate in 50% methanol and then with lead citrate. The grids were viewed in the Philips 410 electron microscope (FEI, Hillsboro, Oregon, United States) at 80 kV and images were recorded on Kodak (Rochester, New York, United States) SO-163 film.
TUNEL staining.
Pancreas tissue for TUNEL staining was fixed in 4% paraformaldehyde, sectioned at 5 μm thick, with TUNEL staining performed by Histoserve. Visualization of TUNEL-positive cells was done using New Fushcin dye. Each of the results for the two wild-type animals (mean counts of 0.33 and 0.25 per field) was compared to each of the two results for the pq/pq animals (mean counts of 7.2 and 4.9 per field) in a Bonferroni multiple-comparison test (pp. 255–262 of [38]). Because the distributions had significantly different standard deviations, the test was repeated with the non-parametric Kruskal-Wallis test followed by Dunn's post test [38]. All four pair-wise comparisons were statistically significant at p < 0.01.
Cell preparation and flow cytometry.
Bone marrow cells were obtained by injecting PBS containing 5% FCS through surgically extracted femurs. Splenic cells and thymocytes were obtained by dissociation of the tissue with a plastic mesh and rubber stopper from a 3-ml syringe. All cells were treated with red blood cell lysis solution. For flow cytometric analysis, cells were blocked with normal mouse serum and Fc block for 10 min, then stained with FITC-, PE-, CyChrome-, or biotin-conjugated antibodies for 30 min. Biotin-conjugated antibodies were detected with a secondary stain: streptavidin-APC. Analysis was performed on a FACScan (Becton-Dickinson, Palo Alto, California, United States) using Flowjo software. The following monoclonal antibodies were obtained from BD PharMingen (San Diego, California, United States): CD3, CD4, CD8, B220, CD19, CD43, CD24, CD11b, GR-1, Ter119, DX5, and IgM. Anti-IgD was purchased from Southern Biotechnology (Birmingham, Alabama, United States).
Genotyping.
PCR reactions used 1× Genamp PCR buffer, 2.5 mM MgCl2, 0.2 mM dNTPs, 0.2 nM each primer, and 1 unit Amplitaq (Applied Biosystems, Foster City, California, United States). PCR conditions were 5 min. denaturation at 93 °C followed by 35 cycles of 93 °C for 30 sec, 58 °C for 30 sec, and 72 °C for 30 sec. BAC primers designed to flanking arm sequences were 3199R (CGCTGCAAAACGCTGACGGAACAGTAG) and 3670R (CTCAGCGTATGGTTGTCGCCGGATGTAT), and 11016F (ATGGGCAAATATTATACGCAGGCGACAAG) and 11595R (CGTATTAGCGGCCGCAAATTTATTAGAGCA). Primers within the pequeño deletion, but outside of BAC sequences were 77kbF (CTTGCCTCATCAATGCTAGG) and 77kbR (GGATAAGTCTATCTGACTTC). Primers specific to transgene insertion were P3p39 (CTGGAGCCTGTGGACTTGGAT) and TVA Rout2 (CAGTGATCAGCATCCACATGC).
For primers used to characterize the pequeño genomic deletion, see Table S1.
Supporting Information
(A) Flow cytometric analysis was conducted by gating on granulocytes using forward versus side light scatter and GR-1and CD11b staining in the bone marrow. Increased percentage of neutrophils in the bone marrow of pq/pq mice is due to elevated percentage of GR-1lo CD11b+ immature neutrophils.
(696 KB PDF)
(63 KB DOC)
(54 KB PDF)
Accession Numbers
The OMIM (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIM) accession numbers for the genes and gene products discussed in this article are CF (602421) and SDS (260400).
The MGI (http://www.informatics.jax.org/) accession number for Serpini2 is 1915181.
The Ensembl (http://www.ensembl.org) accession numbers for the genes discussed in this article are PRSS1 (ENSG00000173636), SPINK1 (ENSG00000164266), and E-62199.1/E-38943.1 (ENSMUSG00000058653).
The UniGene (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=unigene) accession number of the human ortholog of the mouse gene Serpini2 discussed in this article is Hs.376728.
Acknowledgments
We gratefully acknowledge Karen Stoos and Jennifer Puck for providing the BAC genotyping primer sequences, Ana Venegas and Li Jun Yu for providing technical assistance, and Mike Cichanowski and Julia Fekecs for graphics assistance. Animal procedures were performed in accordance with protocol G94–7 from the National Human Genome Research Institute,National Institutes of Health Institutional Review Board. This research was supported by the Intramural Research program of the National Human Genome Research Institute, National Institutes of Health.
Abbreviations
- BAC
bacterial artificial chromosome
- CF
cystic fibrosis
- FISH
fluorescent in situ hybridization
- PI
pancreatic insufficiency
- pq,
pequeño;
- SDS
Shwachman-Diamond Syndrome
- TUNEL
terminal deoxynucleotidyl transferase biotin–dUTP nick end labeling
- ZG
zymogen granule
Footnotes
Competing interests. The authors have declared that no competing interests exist.
Author contributions. SKL, LGB, PLS, and WJP conceived and designed the experiments. SKL, JLC, AI, EP, AC, PMZ, and MAB performed the experiments. SKL, JLC, AI, EP, AC, PMZ, MAB, LGB, PLS, and WJP analyzed the data. SKL, JLC, EP, AC, PMZ, MAB, and PLS contributed reagents/materials/analysis tools. SKL, JLC, PMZ, MAB, LGB, PLS, and WJP wrote the paper.
A previous version of this article appeared as an Early Online Release on August 18, 2005 (DOI: 10.1371/journal.pgen.0010038.eor).
References
- Meisler MH. Insertional mutation of ‘classical' and novel genes in transgenic mice. Trends Genet. 1992;8:341–344. doi: 10.1016/0168-9525(92)90278-c. [DOI] [PubMed] [Google Scholar]
- Rowntree RK, Harris A. The phenotypic consequences of CFTR mutations. Ann Hum Genet. 2003;67:471–485. doi: 10.1046/j.1469-1809.2003.00028.x. [DOI] [PubMed] [Google Scholar]
- Dror Y, Freedman MH. Shwachman-diamond syndrome. Br J Haematol. 2002;118:701–713. doi: 10.1046/j.1365-2141.2002.03585.x. [DOI] [PubMed] [Google Scholar]
- Reid LE, Walker NI. Acinar cell apoptosis and the origin of tubular complexes in caerulein-induced pancreatitis. Int J Exp Pathol. 1999;80:205–215. doi: 10.1046/j.1365-2613.1999.00116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamoto N, Ashizawa N, Niigaki M, Kaji T, Katsube T, et al. Morphological changes in the rat exocrine pancreas after pancreatic duct ligation. Histol Histopathol. 2002;17:1033–1041. doi: 10.14670/HH-17.1033. [DOI] [PubMed] [Google Scholar]
- Durie PR, Kent G, Phillips MJ, Ackerley CA. Characteristic multiorgan pathology of cystic fibrosis in a long-living cystic fibrosis transmembrane regulator knockout murine model. Am J Pathol. 2004;164:1481–1493. doi: 10.1016/S0002-9440(10)63234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L, Loftus SK, Incao A, Chen A, Pavan WJ. Complementation of melanocyte development in SOX10 mutant neural crest using lineage-directed gene transfer. Dev Dyn. 2004;229:54–62. doi: 10.1002/dvdy.10468. [DOI] [PubMed] [Google Scholar]
- Cummings MC, Winterford CM, Walker NI. Apoptosis. In: Sternberg SS, editor. Histology for pathologists, 2nd ed. Philadelphia: Lippincott-Raven; 1997. pp. 3–21. [Google Scholar]
- Ozaki K, Nagata M, Suzuki M, Fujiwara T, Miyoshi Y, et al. Isolation and characterization of a novel human pancreas-specific gene, pancpin, that is down-regulated in pancreatic cancer cells. Genes Chromosomes Cancer. 1998;22:179–185. [PubMed] [Google Scholar]
- Chang WS, Chang NT, Lin SC, Wu CW, Wu FY. Tissue-specific cancer-related serpin gene cluster at human chromosome band 3q26. Genes Chromosomes Cancer. 2000;29:240–255. [PubMed] [Google Scholar]
- Meffre E, Casellas R, Nussenzweig MC. Antibody regulation of B cell development. Nat Immunol. 2000;1:379–385. doi: 10.1038/80816. [DOI] [PubMed] [Google Scholar]
- Melchers F, ten Boekel E, Seidl T, Kong XC, Yamagami T, et al. Repertoire selection by pre-B-cell receptors and B-cell receptors, and genetic control of B-cell development from immature to mature B cells. Immunol Rev. 2000;175:33–46. [PubMed] [Google Scholar]
- Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao G, Liu YE, Gentz R, Sang QA, Ni J, et al. Suppression of breast cancer growth and metastasis by a serpin myoepithelium-derived serine proteinase inhibitor expressed in the mammary myoepithelial cells. Proc Natl Acad Sci U S A. 1999;96:3700–3705. doi: 10.1073/pnas.96.7.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarff KL, Ung KS, Nandurkar H, Crack PJ, Bird CH, et al. Targeted disruption of SPI3/Serpinb6 does not result in developmental or growth defects, leukocyte dysfunction, or susceptibility to stroke. Mol Cell Biol. 2004;24:4075–4082. doi: 10.1128/MCB.24.9.4075-4082.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman GA, Bird PI, Carrell RW, Church FC, Coughlin PB, et al. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J Biol Chem. 2001;276:33293–33296. doi: 10.1074/jbc.R100016200. [DOI] [PubMed] [Google Scholar]
- van Gent D, Sharp P, Morgan K, Kalsheker N. Serpins: Structure, function and molecular evolution. Int J Biochem Cell Biol. 2003;35:1536–1547. doi: 10.1016/s1357-2725(03)00134-1. [DOI] [PubMed] [Google Scholar]
- Huntington JA, Carrell RW. The serpins: Nature's molecular mousetraps. Sci Prog. 2001;84:125–136. doi: 10.3184/003685001783239032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Cronshagen U, Kern HF. A novel pancreas-specific serpin (ZG-46p) localizes to the soluble and membrane fraction of the Golgi complex and the zymogen granules of acinar cells. Eur J Cell Biol. 1997;73:205–214. [PubMed] [Google Scholar]
- Schmidt K, Dartsch H, Linder D, Kern HF, Kleene R. A submembranous matrix of proteoglycans on zymogen granule membranes is involved in granule formation in rat pancreatic acinar cells. J Cell Sci. 2000;113:2233–2242. doi: 10.1242/jcs.113.12.2233. [DOI] [PubMed] [Google Scholar]
- Banks WJ. Applied veterinary histology. Baltimore: Williams and Wilkins; 1986. 527. p. [Google Scholar]
- Whitcomb DC, Gorry MC, Preston RA, Furey W, Sossenheimer MJ, et al. Hereditary pancreatitis is caused by a mutation in the cationic trypsinogen gene. Nat Genet. 1996;14:141–145. doi: 10.1038/ng1096-141. [DOI] [PubMed] [Google Scholar]
- Witt H, Luck W, Hennies HC, Classen M, Kage A, et al. Mutations in the gene encoding the serine protease inhibitor, Kazal type 1 are associated with chronic pancreatitis. Nat Genet. 2000;25:213–216. doi: 10.1038/76088. [DOI] [PubMed] [Google Scholar]
- Bhatia M, Wallig MA, Hofbauer B, Lee HS, Frossard JL, et al. Induction of apoptosis in pancreatic acinar cells reduces the severity of acute pancreatitis. Biochem Biophys Res Commun. 1998;246:476–483. doi: 10.1006/bbrc.1998.8519. [DOI] [PubMed] [Google Scholar]
- Kaiser AM, Saluja AK, Sengupta A, Saluja M, Steer ML. Relationship between severity, necrosis, and apoptosis in five models of experimental acute pancreatitis. Am J Physiol. 1995;269:C1295–C1304. doi: 10.1152/ajpcell.1995.269.5.C1295. [DOI] [PubMed] [Google Scholar]
- Gukovskaya AS, Gukovsky I, Jung Y, Mouria M, Pandol SJ. Cholecystokinin induces caspase activation and mitochondrial dysfunction in pancreatic acinar cells. Roles in cell injury processes of pancreatitis. J Biol Chem. 2002;277:22595–22604. doi: 10.1074/jbc.M202929200. [DOI] [PubMed] [Google Scholar]
- Gukovskaya AS, Perkins P, Zaninovic V, Sandoval D, Rutherford R, et al. Mechanisms of cell death after pancreatic duct obstruction in the opossum and the rat. Gastroenterology. 1996;110:875–884. doi: 10.1053/gast.1996.v110.pm8608898. [DOI] [PubMed] [Google Scholar]
- Vacchio MS, Ashwell JD. Glucocorticoids and thymocyte development. Semin Immunol. 2000;12:475–485. doi: 10.1006/smim.2000.0265. [DOI] [PubMed] [Google Scholar]
- Fantuzzi G, Faggioni R. Leptin in the regulation of immunity, inflammation, and hematopoiesis. J Leukoc Biol. 2000;68:437–446. [PubMed] [Google Scholar]
- Laakko T, Schwartz RC, Fraker PJ. IL-7-mediated protection of pro and pre-B cells from the adverse effects of corticosterone. Cell Immunol. 2002;220:39–50. doi: 10.1016/s0008-8749(03)00023-6. [DOI] [PubMed] [Google Scholar]
- Pazirandeh A, Xue Y, Prestegaard T, Jondal M, Okret S. Effects of altered glucocorticoid sensitivity in the T cell lineage on thymocyte and T cell homeostasis. FASEB J. 2002;16:727–729. doi: 10.1096/fj.01-0891fje. [DOI] [PubMed] [Google Scholar]
- Boocock GR, Morrison JA, Popovic M, Richards N, Ellis L, et al. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat Genet. 2003;33:97–101. doi: 10.1038/ng1062. [DOI] [PubMed] [Google Scholar]
- Nakashima E, Mabuchi A, Makita Y, Masuno M, Ohashi H, et al. Novel SBDS mutations caused by gene conversion in Japanese patients with Shwachman-Diamond syndrome. Hum Genet. 2004;114:345–348. doi: 10.1007/s00439-004-1081-2. [DOI] [PubMed] [Google Scholar]
- Woloszynek JR, Rothbaum RJ, Rawls AS, Minx PJ, Wilson RK, et al. Mutations of the SBDS gene are present in most patients with Shwachman-Diamond syndrome. Blood. 2004;104:3588–3590. doi: 10.1182/blood-2004-04-1516. [DOI] [PubMed] [Google Scholar]
- Lundsteen C, Lind AM. A test of a climate room for preparation of chromosome slides. Clin Genet. 1985;28:260–262. doi: 10.1111/j.1399-0004.1985.tb00397.x. [DOI] [PubMed] [Google Scholar]
- Pinkel D, Straume T, Gray W. Cytogenetic analysis using quantitative, high-sensitivity fluorescence hybridization. Proc Natl Acad Sci U S A. 1986;83:2934–2938. doi: 10.1073/pnas.83.9.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichter P, Cremer T, Borden J, Manuelidis L, Ward DC. Delineation of individual human chromosomes in metaphase and interphase cells by in situ suppression hybridization using recombinant DNA libraries. Hum Genet. 1988;80:224–234. doi: 10.1007/BF01790090. [DOI] [PubMed] [Google Scholar]
- Motulsky H. Intuitive Biostatistics. New York: Oxford University Press; 1995. 386. p. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Flow cytometric analysis was conducted by gating on granulocytes using forward versus side light scatter and GR-1and CD11b staining in the bone marrow. Increased percentage of neutrophils in the bone marrow of pq/pq mice is due to elevated percentage of GR-1lo CD11b+ immature neutrophils.
(696 KB PDF)
(63 KB DOC)
(54 KB PDF)