Abstract
Retroviral reverse transcriptases contain a DNA polymerase activity that can copy an RNA or DNA template and an RNase H activity that degrades the viral RNA genome during reverse transcription. RNase H makes both specific and nonspecific cleavages; specific cleavages are used to generate and remove the polypurine tract primer used for plus-strand DNA synthesis and to remove the tRNA primer used for minus-strand DNA synthesis. We generated mutations in an HIV-1-based vector to change amino acids in the RNase H domain that contact either the RNA and DNA strands. Some of these mutations affected the initiation of DNA synthesis, demonstrating an interdependence of the polymerase and RNase H activities of HIV-1 reverse transcription during viral DNA synthesis. The ends of the linear DNA form of the HIV-1 genome are defined by the specific RNase H cleavages that remove the plus- and minus-strand primers; these ends can be joined to form two-long-terminal repeat circles. Analysis of two-long-terminal repeat circle junctions showed that mutations in the RNase H domain affect the specificity of RNase H cleavage.
HIV-1 reverse transcriptase (RT) is the virally encoded enzyme that converts the single-stranded RNA genome found in virions into double-stranded DNA. The conversion of the RNA genome into DNA is accomplished through the collaboration of the two enzymatic activities of RT: a DNA polymerase that can use either RNA or DNA as a template and an RNase H that cleaves RNA if (and only if) it is present in an RNA⋅DNA duplex (reviewed in refs. 1–3). Both polymerase and RNase H activities are required for the conversion of the RNA genome into double-stranded DNA; mutations that inactivate either the polymerase or RNase H block viral replication (4–7).
In vitro assays showed that polymerase and RNase H activities of HIV-1 RT are interdependent (8–13). Both the polymerase and RNase H domains of HIV-1 RT simultaneously contact the nucleic acid substrate and contribute to the binding (and proper positioning) of the nucleic acid. In in vitro assays, mutations in the RNase H domain can affect polymerase activity (and vice versa). We found that reducing RNase H activity by cotransfecting a mixture of wild-type HIV-1 vector DNA and DNA encoding HIV-1 vectors with mutations in the RNase H active site reduced the efficiency of initiation of DNA synthesis (14). On the basis of these observations, we wanted to determine whether mutating amino acids that make specific contacts with the nucleic acid substrate in the vicinity of the RNase H active site would also affect the initiation of DNA synthesis and/or the specificity of RNase H cleavage.
Reverse transcription requires that RNase H make both specific and nonspecific cleavages. The process of reverse transcription is shown schematically in Fig. 1 (reviewed in refs. 1 and 3). The RNase H cleavages that remove the tRNA primer, and the cleavages that generate and remove the polypurine tract (ppt) primer define the ends of the unintegrated linear viral DNA that is the substrate for the integration reaction. HIV-1 RT removes the tRNA one base from the RNA-DNA junction (15–18); other retroviral RTs remove the entire tRNA (2). Removal of the tRNA primer defines the right end of the viral DNA; the generation and removal of the ppt primer defines the left end of the viral DNA. If RNase H fails to remove either the tRNA or the ppt primers, additional sequences will be present at the ends of the viral DNA. Although retention of the primers would interfere with integration, the presence of the primers would not interfere with the strand-transfer reactions.
Figure 1.
Reverse transcription of viral RNA. (i) Minus-strand DNA synthesis is initiated from a tRNALys-3 primer (gray arrow) annealed to the pbs. (ii) The U5 and R regions of the viral RNA (thin line) are copied into DNA (thick line), and the RNase H activity of RT degrades the viral RNA (the dotted line represents degraded RNA). (iii) Minus-strand DNA can be transferred to the 3′ end of the viral RNA because of the complementarity of the viral RNA R region and the minus-strand DNA. (iv) Minus-strand DNA elongation copies the RNA genome, and the RNase H activity of RT degrades the viral RNA. The ppt is resistant to RNase H cleavage and serves as the primer for plus-strand DNA synthesis. Two specific cleavages by RNase H generate the ppt primer (arrowheads). (v) Plus-strand DNA synthesis is initiated from the ppt and copies the U3, R, and U5 regions of the minus-strand DNA. RNase H removes the tRNA primer 1 base from the RNA/DNA junction and the ppt at the RNA/DNA junction (arrowheads). (vi) Plus-strand transfer occurs using the complementarity of the pbs and the ends of the viral DNA. Extension of the plus- and minus-strands complete the synthesis of the viral DNA. The ribonucleotide (rA) at the 5′ end of the minus strand is derived from the tRNA. HIV-1 RT removes the tRNA one base from the RNA-DNA junction (15–18); other retroviral RTs remove the entire tRNA (2). (vii) The structure of full-length viral DNA.
On the basis of the crystal structure of HIV-1 RT in complex with an RNA-DNA template primer, we generated a series of mutations in the HIV-1 RT RNase H primer grip [a network of amino acids that contacts the primer strand near the RNase H active site (19)] and in amino acids that contact the RNA template near the RNase H active site (Fig. 2). The idea that the RNase H primer grip plays a critical role in how RT interacts with its nucleic acid substrate is supported by data that show that contacts with the DNA primer strand are similar in HIV-1 RT in complexes with RNA-DNA and DNA-DNA template primers (19–21). If these mutations alter the positioning of the nucleic acid relative to the polymerase active site or the RNase H active site, this could, in turn, affect DNA synthesis and/or induce alterations in the RNase H cleavages. Altering RNase H cleavages could affect the proper removal of the tRNA and the generation and removal of the ppt primer. For the mutants tested, the decrease in titer was due primarily to a decrease in the efficiency of the initiation of viral DNA synthesis. We also tested the effects of these mutations on the specificity of RNase H cleavages by monitoring the sequences at the ends of the viral genome (22).
Figure 2.
Stereo diagram showing the structure of the RNase H primer grip. The RNA template strand is shown in turquoise, the DNA primer strand, in purple. The scissile phosphate is designated with a red arrow pointing to the phosphate. The RNase H domain is shown in orange; the connection subdomain is in yellow. Amino acids contacting the nucleic acid are labeled. Amino acids that were not mutated are labeled in black. Amino acids that had minimal (<2-fold) effects on titer when converted to alanine are labeled in green. Amino acids where alanine substitutions had a moderate (>5-fold) or strong effect on titer are labeled in blue. Contacts between individual amino acids and the RNA template are shown in black, contacts with the DNA primer are shown in red. Most of the contacts are shown as dotted lines; contacts with Q475 (which contacts both strands) are shown as solid lines. The nucleic acid strands are numbered relative to the site of cleavage on the RNA strand.
The two-long-terminal-repeat (2-LTR) circles arise from the joining of the ends of unintegrated viral DNA, presumably by host cell ligases (23). The 2-LTR circles derive from the portion of viral linear DNA that fails to integrate; linear DNAs with abnormal ends are not good substrates for integration and are more likely to be converted into circular DNA. Because we are particularly interested in viral DNAs with abnormal ends, this enrichment should be helpful. As a surrogate for the ends of the viral DNA, we analyzed the 2-LTR circle junction sequences derived from cells infected with wild-type virus and viruses with mutations in the RNase H primer grip that displayed moderate (5- to 10-fold) reductions in virus titer by using an HIV-1 vector that undergoes a single replication cycle. Samples derived from cells infected with mutant viruses displaying moderate decreases in virus titer had a significantly larger proportion of aberrant circle junctions than samples derived from the cells infected with the wild-type virus. In vitro analysis showed that some of the mutations affect the specificity of the RNase H cleavage; these cleavage defects can explain the genesis of some of the aberrant 2-LTR circle junctions.
Materials and Methods
Site-Directed Mutagenesis.
The Asp718 to SalI fragment of pNLNgoMIV R−E−.HSA was cloned into the plasmid KS (Stratagene) (14). Mutations in the RNase H primer grip were generated by using the Quick-Change mutagenesis kit (Stratagene) and confirmed by DNA sequencing.
Cells.
Human embryonic kidney cell line 293 was obtained from American Type Culture Collection. Human osteosarcoma cell line HOS was obtained from Richard Schwartz (Michigan State University, Lansing, MI). 293 and HOS cells were maintained as previously described (14).
Determination of Virus Titer.
Virus was produced by pseudotyping the pNLNgoMIVR−E−.HSA vectors with the VSV-g envelope glycoprotein (expressed from pHCMV-g; obtained from Jane Burns, University of California, San Diego). The vector undergoes only a single cycle of replication. Virus titers were determined as previously described (14).
Analysis of 2-LTR Circle Junctions.
293 cells were transfected with 5 μg of pNLNgoMIVR−E−.HSA vector and 3 μg of pHCMV-g by the calcium phosphate method. The 48-h supernatants were harvested and used to infect HOS cells plated at 1 × 106 cells per 100-mm-diameter plate on the day before infection. The supernatants were left on the cells for 4 h, then fresh medium was added to the cells. Thirty-six hours after infection, the total DNA was isolated by the QIAamp DNA Blood Mini kit (Qiagen, Chatsworth, CA). The 2-LTR circle junctions were amplified in 100-μl reactions by using an upstream oligonucleotide that anneals near the RU5 junctions and a downstream oligonucleotide that anneals to the U3 region of the LTR. The sequence of the upstream primer was 5′-CGATGAATTCGCTAACTAGGGAACCCACTGCT-3′; the sequence of the downstream primer was 5′-GCCATTCTAGAGTTCTCTCCTTTATTGGCCTC-3′. Each PCR sample contained 9 μl of total DNA, 90 μl of Platinum PCR Supermix (Invitrogen), and 0.5 μl of upstream and downstream primers (final concentration of 100 nM for each primer). The size of the PCR product was approximately 350 bp, and the fragment was flanked by EcoRI and XbaI sites that were introduced by the PCR primers. The PCR product was digested with EcoRI and XbaI and cloned into the plasmid SK (Stratagene). Clones containing 2-LTR circle junctions were analyzed by digestion with restriction enzymes and DNA sequencing.
Determination of DNA Copy Number by Real-Time PCR.
Real-time PCR was used to determine the effects of mutations on DNA synthesis, as previously described (14).
Results and Discussion
Effects of RNase H Mutations on Viral Titer.
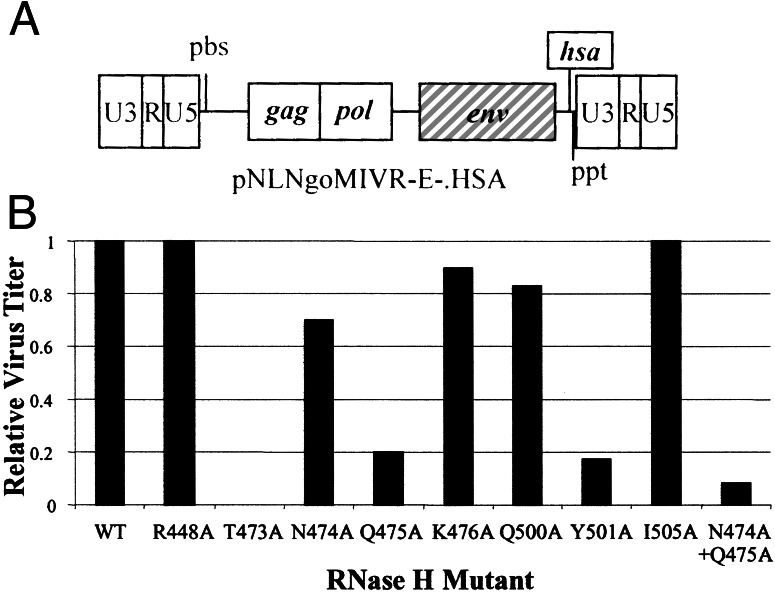
Mutations were made that substitute alanines for amino acids in the RNase H primer grip (T473, I505, K476, Q475, Y501) and in the amino acids that contact the RNA template near the RNase H active site (R448, N474, Q475, Q500). One mutant is in both groups: Q475 contacts both the RNA and DNA strands (Fig. 2). The mutations were introduced into an HIV-1-based vector system that was limited to single round of replication (Fig. 3A; ref. 14) and tested for their effects on viral titer (Fig. 3B). T473A abolished replication, R448A and I505A had no discernible effect on titer; and N474A, K476A, and Q500A reduced the titer less than 2-fold. Q475A, Y501A, and the N474A + Q475A double mutation reduced the titer 5- to 10-fold.
Figure 3.
(A) The HIV-1 vector pNLNgoMIVR−E−.HSA expresses the murine cell surface marker CD24 (heat-stable antigen, HSA) from the nef ORF. The env and the vpr genes have been inactivated. The drawing is not to scale. (B) The effect of mutations in the RNase H domain on virus titer. The relative infectivity of the mutants, normalized to the p24 concentration, is shown on the y axis and the mutants are shown on the x axis.
All of the single mutations that had significant effects on replication (T473A, Q475A, and Y501A) are located in the RNase H primer grip and contact the DNA primer relatively close to the RNase H active site at positions −4 or −5. [The numbering of the DNA positions relative to the RNA is affected by mispairing in the ppt (19).] However, Q475 also contacts the RNA strand (at −1 and −2). In each case, the alanine substitution changes the nature of the amino acid in a fashion expected to disrupt the interaction with the nucleic acid. In contrast, alanine substitutions at R448, N474, and Q500 had little or no effect on viral titer. These amino acids contact the RNA template, rather than the DNA primer. These data suggest that, in the vicinity of the RNase H active site, the DNA contacts may be more important for infectivity than the RNA contacts. I505 contacts the phosphate between −5 and −6 on the DNA primer; however, the I505A substitution may not have perturbed the nature and size of the amino acid sufficiently to produce a significant effect on viral replication. K476 contacts the phosphate between −4 and −5 on the DNA primer. The K476A substitution did change the nature of the amino acid but had only a small (less than 2-fold) effect on replication.
Effects of the Y501A Mutation and the N474A + Q475A Double Mutation on Viral DNA Synthesis.
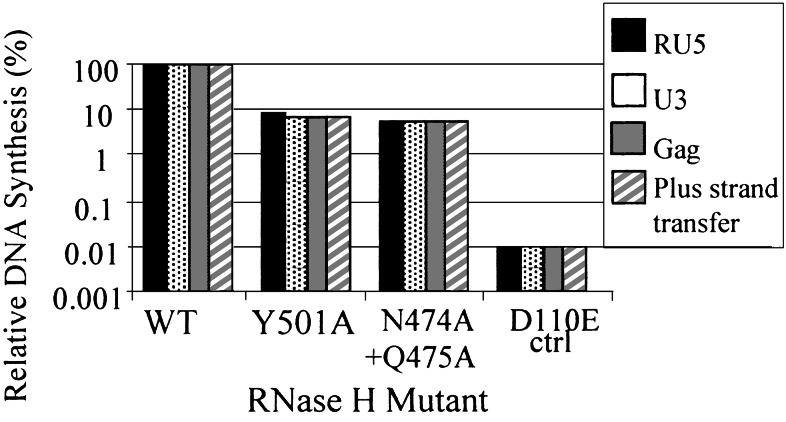
Viral DNA synthesis was monitored in cells infected with mutants that had a significant effect on viral titer (Y501A and N474A + Q475A). Analysis of the viral DNA showed that Y501A and N474A +Q475A reduced the efficiency of DNA synthesis (Fig. 4). During the first step of reverse transcription, the R and U5 regions of the viral RNA are copied into minus-strand DNA. To monitor the initiation of DNA synthesis, we measured the amount of RU5 DNA synthesized by real-time PCR (Fig. 4). The effects of the primer grip mutations on the initiation of DNA synthesis were similar to the effects of the mutations on virus titer (Fig. 3B). To measure the first-strand transfer, the level of RU5 DNA was compared with the level of U3 DNA (see Fig. 1). The Y501A mutation caused a small reduction in the relative amount of U3 DNA compared with the wild-type virus; this suggests that the mutation may have a modest effect on first-strand transfer. Measuring the level of gag specific DNA sequences monitors elongation; measuring the level of DNA spanning the primer-binding site (pbs) region measures plus-strand DNA transfer. Y501A had no measurable effects on minus-strand elongation or on second-strand transfer. The N474A + Q475A double mutant had no measurable effect on first- or second-strand transfer (Fig. 4).
Figure 4.
Viral DNA synthesis by RNase H mutants. The amount of viral DNA synthesized by the RNase H primer grip mutants relative to wild-type is shown on the y axis. The virus used to infect cells is indicated on the x axis the different steps of reverse transcription that were monitored are shown by the differently shaded bars, as designated in the figure key. The amount of virus used for the infections was measured by using a p24 ELISA. The copy numbers of different products measured for the different steps of reverse transcription for wild-type virus were 6 × 105 copies of RU5 specific DNA, 5 × 105 copies of U3 specific DNA, 4 × 105 copies of gag-specific DNA, and 2 × 105 copies of DNA specific for plus-strand transfer.
In vitro analysis suggested that the initiation of viral DNA synthesis is a difficult step for HIV-1 RT (24, 25). Previous experiments in cell culture with viruses derived by transfecting mixtures of a wild-type vector and a vector containing the RNase H active site mutation E478Q mutation showed that decreasing the level of RNase H activity significantly reduced the initiation of viral DNA synthesis (14). Mutations in the RNase H domain that contact the RNA or DNA strands could perturb the interactions of RT with its nucleic acid substrate, which could affect both RNase H and polymerase activity. Taken together, the available data suggest that the initiation of HIV-1 viral DNA synthesis is a difficult step in vivo as well as in vitro. Minus-strand DNA synthesis is initiated on an RNA⋅RNA duplex. Because an RNA⋅RNA duplex will assume an A-form structure, it may be difficult for the enzyme to bend an RNA⋅RNA duplex to form a structure similar to that found in DNA⋅DNA and RNA⋅DNA duplexes bound to HIV-1 RT (19–21). Bending an RNA⋅RNA duplex may require contacts in both the polymerase and RNase H domains; loss of contacts in either domain could affect the initiation of reverse transcription. Mutations that reduced the initiation of viral DNA synthesis in vivo (Y501A and N474A + Q475A) were tested to determine whether they would reduce the initiation of DNA synthesis in vitro using a 32P-end-labeled tRNALys-3 primer annealed to a 105-nt pbs RNA derived from the HXB2 strain of HIV-1. No measurable effect was seen however the in vitro assays were done in the absence of nucleocapsid and may not have been sensitive enough to show a defect (22).
Analysis of 2-LTR Circle Junctions As a Surrogate for the Ends of the Viral DNA.
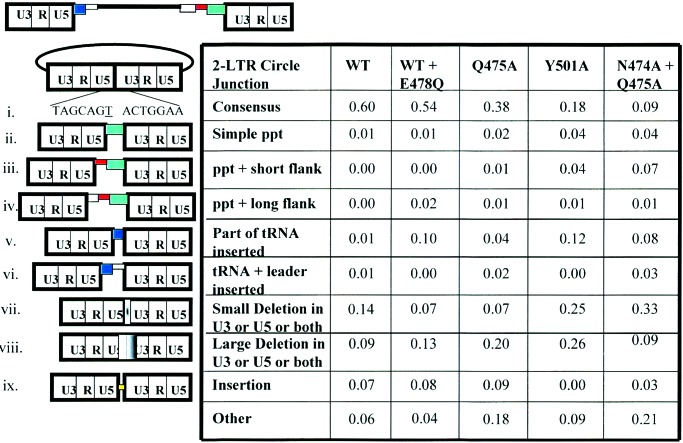
Approximately 100 clones were sequenced from cells infected with the wild-type vector, for a vector with reduced RNase H activity (produced by cotransfection of wild-type vector DNA and vector DNA encoding the RNase H active site mutation E478Q), and for the RNase H primer grip mutants displaying a 5- to 10-fold decrease in titer. For each experiment, clones were derived from at least two independent PCR reactions. In all cases, the independent PCR reactions gave similar sets of aberrant 2-LTR circle junctions. Of the 2-LTR circle junctions derived from infection with the wild-type vector, the fraction that had the consensus sequence was 0.60. This number is similar to results reported previously for HIV-1 where the fraction of correct 2-LTR circle junctions was reported to be 0.59 (10/17, ref. 26) and 0.85 (22/26, ref. 8). A similar frequency of correct 2-LTR junctions was also reported for wild-type Moloney murine leukemia virus (70 of 91, ref. 27); our results with the HIV-1 vector system are similar to the results obtained using replication competent retroviruses.
We first measured the effects of reducing RNase H activity on the sequences at the ends of the viral DNA. Alterations at the circle junctions could be caused either by a reduction in the level of RNase H activity or by changes in the specificity of RNase H cleavage. To distinguish between these possibilities, we determined the effects of decreasing RNase H activity by cotransfecting a mixture of wild-type vector DNA and vector DNA encoding RT with the RNase H active site mutation E478Q. Reducing the level of RNase H activity by approximately 90% (transfecting a 1:9 mixture of wild-type and mutant DNA) reduced the titer about 5-fold (14), similar to the reduction in titer caused by the RNase H primer grip mutations that were chosen for analysis. The initiation of DNA synthesis was decreased in the virions with reduced RNase H activity (14), which is similar to the effect on initiation by the Y501A and N474A + Q475 mutants. Reducing RNase H activity approximately 10-fold had a relatively small effect on the fraction of consensus circle junctions (Fig. 5, line i); however, there was a shift in the pattern of aberrant forms relative to wild-type virus. There was a significant increase in the number of tRNA sequences seen at the circle junction (Fig. 5, line v), suggesting that tRNA removal is sensitive to the overall level of RNase H activity. However, reducing the level of RNase H activity did not significantly affect the number of ppt sequences inserted at the 2-LTR circle junction.
Figure 5.
Frequency of different classes of 2-LTR circle junctions. 2-LTR circle junctions were analyzed from cells infected with the wild-type vector (WT), vector with reduced RNase H activity (WT + E478Q), and vectors containing the RNase H primer grip mutations Q475A, Y501A, and N474A + Q475A. At the top is a schematic diagram of the unintegrated viral DNA. The pbs is shown in navy blue, the viral leader sequence 3′ of the pbs is shown in light blue, the region 5′ of the ppt is shown in gray, the U-tract is red, and the ppt is in green. The drawing is not to scale. (i) The consensus 2-LTR circle junction arising from the ligation of the ends of unintegrated viral DNA. The underlined T is derived from the ribonucleotide A at the 3′ end of the tRNALys-3 primer used for minus-strand DNA synthesis. (ii) “Simple” ppt insertions (green box) that contain part or all of the ppt but no upstream flanking sequences. This class of defective circle junctions may also contain deletions in the U5 region. (iii) Insertion of the ppt (green box) along with short flanking sequences (adjacent red box) immediately upstream of the ppt. These flanking sequences are 1–5 bp in length and are probably caused by improper cleavages by RNase H (see Fig. 6 and Results and Discussion). (iv) Insertion of the ppt with long flanking sequences (gray box) upstream of the ppt. This class of inserts may also include deletions in the U5 region. (v) Insertions of part of the tRNA Lys-3 sequences (blue box) at the 2-LTR circle junction. Only a portion of the tRNA is reverse transcribed and inserted; this portion corresponds to the tRNA sequences that anneal to the pbs. This class of defective circle junctions may also include deletions in the U3 region. (vi) Insertion of tRNA (navy blue box) and viral leader sequence from downstream of the pbs (light blue box). During reverse transcription, plus-strand DNA synthesis is initiated from the ppt primer and copies both the minus-strand DNA and the portion of the tRNA primer that hybridizes to the pbs (Fig. 1). Normally, this plus strand is transferred to the minus strand after the tRNA is removed (see Fig. 1). However, if RNase H fails to remove the tRNA primer before the plus strand is extended a second time to complete the synthesis of viral DNA, the tRNA can be copied a second time. If this happens, the DNA could undergo another strand-transfer reaction, and RT could then copy the adjacent sequences on the leader, which could be captured at the circle junction. This class of defective circle junctions may also include deletions in the U3 region. (vii) Small deletions (1–5 bp) at the circle junction in the U3, U5, or both U3 and U5 (white to black gradation). (viii) Large deletions in the U5, the U3 or both U5 and U3 (white to black box overlapping the circle junction). (ix) Insertions (yellow box) at the 2-LTR circle junction that do not come from either the ppt or the tRNA primer. (x) Deletions with insertions that do not derive from the ppt or the tRNA primer.
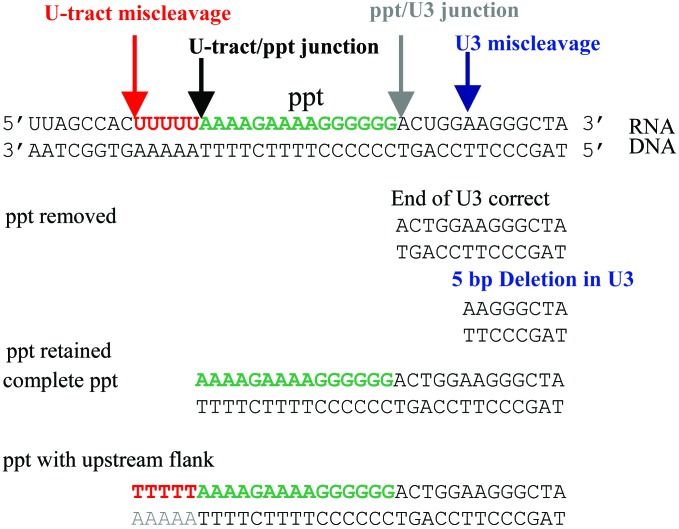
Two specific cleavages are required to generate the normal ppt primer; a third cleavage is required to remove the primer. Reducing the RNase H activity could interfere with any or all of these cleavages. There was no obvious difference in the number of simple ppt sequences (i.e., ppt-derived sequences without any flanking sequences) inserted at the 2-LTR circle junction when RNase H activity was reduced approximately 10-fold; this result suggests that a minimal amount of RNase H activity is sufficient to accurately remove the ppt primer. A simple ppt insert could also be generated if the enzyme failed to cleave at the ppt/U3 junction but did cleave at the junction of the ppt and a run of uracils immediately upstream of the ppt (U-tract) and then used the 3′ end of the U-tract RNA as the plus-strand primer (Fig. 6). That there was no increase in the number of simple ppt inserts suggests that these events do not occur when the RNase H activity is reduced.
Figure 6.
Miscleavages by RNase H lead to insertions of short flanking sequences 5′ of the ppt and to small deletions in the U3 region. (Top) The RNA⋅DNA duplex containing the ppt (green lettering). Wild-type RT cleaves at the U-tract/ppt junction (black arrow) and at the ppt/U3 junction (gray arrow) to generate the normal ppt primer used for plus-strand DNA synthesis. Miscleavage (orange arrow) in the U-tract (orange lettering) can lead to the insertion of flanking sequences if the ppt is retained. Miscleavage in U3 (blue arrow) generates an incorrect ppt primer, which, if it is removed by RNase H, creates a 5-bp deletion in U3.
Reducing the level of RNase H activity did lead to the generation of ppt inserts with long (>100 bp) flanking sequences (Fig. 5). These inserts could arise in two ways: (i) by the initiation of plus-strand DNA synthesis on some upstream RNA primer (not the ppt), which might happen if the cleavages that normally lead to the generation of the ppt primer did not occur; or (ii) by plus-strand initiation from a long ppt primer, which could arise by a failure to cleave at the U-tract/ppt junction. Taken together, the tRNA and ppt insertion data suggest that reducing the level of RNase H activity affects the ability of the enzyme to make some of the specific cleavages it normally makes but does not cause a significant increase in aberrant cleavages near the normal cleavage sites used to generate the ppt primer.
The Y501A and N474A + Q475A RNase H Primer Grip Mutants Have Altered RNase H Cleavage Specificity.
The circle junctions derived from infection with vectors containing the mutant enzymes had more inserts containing ppt sequences than did the circle junctions derived from infection with the wild-type vector or the phenotypically mixed vector with reduced RNase H activity [WT + E478Q (Fig. 5, lines ii–iv)]. This comparison suggests that the mutants have difficulty removing the ppt primer that is not simply a matter of having reduced RNase H activity. In vitro analysis has also shown that the mutant enzymes were less efficient than the wild-type enzyme in their ability to remove a ppt primer from a model substrate; in an in vitro ppt primer removal assay, the Y501A mutant was particularly deficient (22). The RNase H primer grip mutants also generated 2-LTR circle junctions with ppt inserts containing a short flanking sequence. Inserts of this type were not seen in the circle junctions derived from infections with the wild-type vector or the vector with reduced RNase H activity (WT + E478Q).
Changes in the Cleavage Specificity of the Primer Grip Mutants in the U-Tract and U3 Explain Many of the Aberrant 2-LTR Circle Junctions.
Miscleavage within the U-tract or just 5′ of the U-tract would, if the resulting ppt primer was retained, lead to an insertion at the circle junction containing part or all of the U-tract [which would, in the circle junction, appear as a T-tract (Fig. 6)]. Insertion of ppt sequences with a short flanking sequence, including part or all of the T-tract, was not observed in circle junctions derived from infections with the wild-type vector or the infections from the vector stock with reduced RNase H activity (WT + E478Q) (Fig. 5). However, this type of insertion was observed once for samples from infection with Q475A, and several times with samples from Y501A and N474A + Q475A. The fact that insertions of this type were not seen when the RNase H activity was reduced suggests they are caused by an alteration in RNase H specificity and not by an alteration in the level of activity.
If there was an alteration in the specificity of cleavage such that RNase H would cleave within U3 instead of at the ppt/U3 junction, and the aberrant ppt primer was removed, a small deletion would be generated in U3 (Fig. 5, line vii, and Fig. 6). If the mechanism for generating the U3 deletions is miscleavage in U3, there should be no corresponding increase in U5 deletions. Most other mechanisms that would lead to an increase in U3 deletions (a failure to complete DNA synthesis for example) would probably cause deletions in both U3 and U5. In the circle junctions derived from infection with the wild-type virus, there were more junctions with a 1-bp deletion in U5 than in U3 (Table 1). There were a modest number of 2- to 5-bp deletions; however, for the wild-type vector, the frequency of 2- to 5-bp deletions was approximately the same in U3 and U5. Reducing the level of RNase H activity may have caused a slight increase in the 2- to 5-bp deletions in U3; if so, the effect was small. By contrast, both the Y501A mutation and the N474A + Q475A double mutation caused a dramatic increase in the number of 2- to 5-bp deletions in U3; neither caused a corresponding increase in the 2- to 5-bp deletions in U5. If anything, the Q475A mutation decreased the number of 1- and 2- to 5-bp deletions in U3; the Q475A mutation has more modest effects both on viral titer and on the number of consensus circle junctions than the other two mutants (Y501A and N474A + Q475A)
Table 1.
Fraction of small deletions in U3, U5, or both U3 and U5 at the 2-LTR circle junction
| Size of deletion, bp | Wild type
|
Reduced RNase
H
|
Q475A
|
Y501A
|
N474A +
Q475A
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| U5 | U3 | U5 | U3 | U5 | U3 | U5 | U3 | U5 | U3 | |
| 1 | 0.07 | 0.01 | 0.01 | 0.01 | 0.03 | 0.04 | 0.01 | 0.04 | 0.01 | |
| 2 | 0.02 | 0.01 | 0.01 | 0.02 | 0.04 | 0.05 | ||||
| 3 | 0.01 | 0.02 | 0.01 | 0.02 | 0.08 | |||||
| 4 | 0.01 | 0.02 | ||||||||
| 5 | 0.07 | 0.02 | 0.08 | |||||||
| Deletion in U5 | 0.01 | 0.01 | 0.07 | 0.02 | ||||||
| and U3, bp | (1) 1 | 1 | (1) 1 | 1 | (2) 1 | 1 | (2) 1 | 1 | ||
| (4) 1 | 4 | |||||||||
The top portion of the table provides additional information about the circle junctions that have short deletions in U3 or U5 at the 2-LTR circle junction. The size of the deletion is shown on the left-most column, and the samples that the clones were derived from are shown in the columns on the right. The frequency of deletions that were not observed (left empty) are <0.01. The bottom portion of the table shows the frequency of samples containing deletions in both U3 and U5. The frequency of each class of deletion is shown in parentheses, and the sizes for each of the U3 and U5 regions are shown.
The mutants have also been tested for their ability to cleave at the ppt/U3 junction in vitro (22). The wild-type enzyme cleaved preferentially at the junction but also produced some cleavages in the adjacent residues. The three mutants were able to cleave at the junction; however, they were less specific than wild-type RT. All of the mutant enzymes cleaved more extensively in the U3 region adjacent to the ppt; if such cleavages in U3 occurred during viral replication, they would generate small deletions in U3 that would be exactly like those we observed in infections with the vector containing Y501A or N474A + Q475A. The Q475A mutation did not generate deletions of this type in our 2-LTR circle junction assay; however, as has already been mentioned, Q475A had a smaller effect on titer and on the circle junction sequences than did Y501A or the N474A + Q475A double mutation. The ppt insertion and U3 deletion data suggest that the Y501A mutation, the N474A + Q475A double mutation, and possibly the Q475A mutation cause miscleavages during viral replication, both at the U-tract/ppt junction and at the ppt/U3 junction. Infection with a virus carrying the N474A + Q475A double mutation clearly gave rise to many more defective 2-LTR circle junctions than did infection with virus carrying only Q475A. This shows that N474A does have a role in determining the specificity of RNase H cleavage. However, the fact that the effect of the Q475A mutation on viral titer and the initiation of DNA synthesis is nearly as great as the N474A + Q475A double mutant also suggests that the role of N474A in these processes is not as important as its role in determining RNase H specificity.
Taken together, the data provide a link that connects the structural analysis of HIV-1 RT to its role in the replication of the viral genome. There are amino acids in the RNase H domain that contact the nucleic acid and, in so doing, contribute to the specificity of RNase H cleavage both in vitro (22) and in vivo (this report). The in vivo analysis of the effects of the mutations in RNase H also reinforces the importance of the interdependence of the two domains of RT. There are considerable in vitro data to support the idea that appropriate interaction between the enzyme and its nucleic acid substrate depends critically on contacts between the nucleic acid and the protein in both the polymerase and RNase H domains. Because of this interdependence, altering amino acids in RNase H domain affects both the initiation of viral DNA synthesis and the specificity of RNase H cleavage in vivo.
Acknowledgments
We thank David Munroe, Claudia Stewart, and Marilyn Powers for DNA sequencing, Louise Finch for FACS analysis, John Coffin for helpful discussions, Anne Arthur for expert editorial assistance, and Hilda Marusiodis for help in preparing the manuscript. Research in S.H.H.'s laboratory was supported by the National Cancer Institute and by the National Institute for General Medical Sciences. Research in E.A.'s laboratory was supported by National Institutes of Health Grants AI 27690 and GM55609, and S.G.S. was supported by a National Institutes of Health–National Institute of Allergy and Infectious Diseases National Research Service Award fellowship (AI 09578).
Abbreviations
- 2-LTR
two long-terminal-repeat
- RT
reverse transcriptase
- pbs
primer-binding site
- ppt
polypurine tract
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Coffin J M, Hughes S H, Varmus H E. Retroviruses. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. [PubMed] [Google Scholar]
- 2.Hughes S H, Arnold E, Hostomsky Z. In: Ribonucleases H. Crouch R J, Toulmé J J, editors. Paris: Les Editions INSERM; 1998. pp. 195–224. [Google Scholar]
- 3.Whitcomb J M, Hughes S H. Annu Rev Cell Biol. 1992;8:275–306. doi: 10.1146/annurev.cb.08.110192.001423. [DOI] [PubMed] [Google Scholar]
- 4.Repaske R, Hartley J W, Kavlick M F, O'Neill R R, Austin J B. J Virol. 1989;63:1460–1464. doi: 10.1128/jvi.63.3.1460-1464.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schatz O, Cromme F V, Naas T, Lindemann D, Mous J, Le Grice S F J. Gene Regulation and AIDS. Houston: Portfolio; 1990. pp. 293–404. [Google Scholar]
- 6.Tisdale M, Schultze T, Larder B A, Moelling K. J Gen Virol. 1991;72:59–66. doi: 10.1099/0022-1317-72-1-59. [DOI] [PubMed] [Google Scholar]
- 7.Telesnitsky A, Goff S P. EMBO J. 1993;12:4433–4438. doi: 10.1002/j.1460-2075.1993.tb06128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyer P L, Ferris A L, Hughes S H. J Virol. 1992;66:1031–1039. doi: 10.1128/jvi.66.2.1031-1039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyer P L, Ferris A L, Hughes S H. J Virol. 1992;66:7533–7537. doi: 10.1128/jvi.66.12.7533-7537.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyer P L, Tantillo C, Ferris A L, Arnold E, Hughes S H. J Mol Biol. 1994;243:472–483. doi: 10.1006/jmbi.1994.1673. [DOI] [PubMed] [Google Scholar]
- 11.Hizi A, McGill C, Hughes S H. Proc Natl Acad Sci USA. 1988;85:1218–1222. doi: 10.1073/pnas.85.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hizi A, Hughes S H, Shaharabany M. Virology. 1990;175:575–580. doi: 10.1016/0042-6822(90)90444-v. [DOI] [PubMed] [Google Scholar]
- 13.Prasad V, Goff S P. Proc Natl Acad Sci USA. 1989;86:2104–3108. doi: 10.1073/pnas.86.9.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Julias J G, Ferris A L, Boyer P L, Hughes S H. J Virol. 2001;75:6537–6546. doi: 10.1128/JVI.75.14.6537-6546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whitcomb J M, Kumar R, Hughes S H. J Virol. 1990;64:4903–4906. doi: 10.1128/jvi.64.10.4903-4906.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furfine E S, Reardon J E. Biochemistry. 1991;30:7041–7046. doi: 10.1021/bi00243a001. [DOI] [PubMed] [Google Scholar]
- 17.Pullen K A, Ishimoto L K, Champoux J J. J Virol. 1992;66:367–373. doi: 10.1128/jvi.66.1.367-373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith J S, Roth M. J Biol Chem. 1992;267:15071–15079. [PubMed] [Google Scholar]
- 19.Sarafianos S G, Das K, Tantillo C, Clark A D, Jr, Ding J, Whitcomb J, Gait M, Boyer P L, Hughes S H, Arnold E. EMBO J. 2001;20:1449–1462. doi: 10.1093/emboj/20.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding J, Hsiou Y, Sarafianos S G, Clark A D, Jr, Jacobo-Molina A, Tantillo C, Arnold E. J Mol Biol. 1998;284:1095–1111. doi: 10.1006/jmbi.1998.2208. [DOI] [PubMed] [Google Scholar]
- 21.Jacobo-Molina A, Ding J, Nanni R G, Clark A D, Jr, Lu X, Tantillo C, Williams R L, Kamer G, Ferris A L, Clark P, et al. Proc Natl Acad Sci USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rausch J W, Miller J T, Lener D, Julias J G, Hughes S H, Le Grice S F J. Biochemistry. 2002;41:4856–4865. doi: 10.1021/bi015970t. [DOI] [PubMed] [Google Scholar]
- 23.Li L, Olvera J M, Yoder K E, Mitchell R S, Butler S L, Lieber M, Martin S L, Bushman F D. EMBO J. 2001;20:3272–3281. doi: 10.1093/emboj/20.12.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Isel C, Westhof E, Massire C, Le Grice S F, Ehresmann B, Ehresman C, Marquet R. EMBO J. 1999;18:1038–1048. doi: 10.1093/emboj/18.4.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Launchy J M, Isel C, Keith G, Le Grice S F, Ehresmann C, Ehresmann B, Marquet R. J Biol Chem. 2000;275:12306–12312. doi: 10.1074/jbc.275.16.12306. [DOI] [PubMed] [Google Scholar]
- 26.Hong T, Drlica K, Pinter A, Murphy E. J Virol. 1991;65:551–555. doi: 10.1128/jvi.65.1.551-555.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gorelick R J, Fu W, Gagliardi T D, Bosche W J, Rein A, Henderson L E, Arthur L O. J Virol. 1999;73:8185–8195. doi: 10.1128/jvi.73.10.8185-8195.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]