Abstract
Understanding molecular mechanisms that control cell fate in the shoot apical meristem is a fundamental question in plant development. Genetic and molecular studies demonstrate that maize KNOTTED1 (KN1) of the TALE (3-aa acid loop extension) class of homeodomain (HD) proteins is involved in shoot apical meristem function. We show that KN1 interacts with knotted interacting protein (KIP), a BEL1-like TALE HD protein. Interaction between KN1 and KIP is mediated by conserved domains in the N termini of both proteins. The KN1 DNA-binding sequence, TGACAG(G/C)T, was biochemically identified, and in vitro DNA-binding assays show that individually KN1 and the HD of KIP bind specifically to this motif with low affinity. The KN1–KIP complex, however, binds specifically to this DNA-binding motif with high affinity, indicating that the association of KN1 and KIP may function in transcriptional regulation.
In plants, development of the aerial shoot is controlled by a group of cells in the shoot apex called the shoot apical meristem (SAM). The SAM is the site where organogenesis is initiated and from which leaves, flowers, or other determinate structures are produced. Because plant development is a continuous process, the SAM must balance the process of differentiation and maintenance to allow growth.
Maize knotted1 (kn1) and the Arabidopsis ortholog SHOOTMERISTEMLESS (STM) are members of the knox family of genes that are required for SAM function (1). The expression of kn1 and STM is restricted to the indeterminate cells of the SAM. Moreover, this pattern of expression is excluded from the zone of the SAM that gives rise to organs (2, 3). Loss-of-function mutations in kn1 and STM produce a shootless phenotype, where the SAM prematurely terminates after initiating cotyledons, demonstrating that kn1 and STM function in meristem maintenance (3–5). In backgrounds where some shoot formation occurs, organ production is reduced, indicating that vegetative and inflorescence meristems are compromised (4, 6–8). In addition to maintenance functions, class 1 knox genes, including kn1, are capable of respecifying cell fates when ectopically expressed in the leaf (1). Studies that support this hypothesis come from analysis of dominant mutations in the maize knox genes, kn1, rough sheath1 (rs1), liguleless3 (lg3), and gnarley1 (gn1) (9–12). Each of these phenotypes is caused by ectopic leaf expression, resulting in proximal leaf cell identities transposed distally into the blade (1, 13).
The knox gene family was the first group of plant genes identified that encodes homeodomain (HD) proteins (14). In animals, HD-containing proteins function as transcription factors that are important regulators of animal cell fate and development (15). The HD is a conserved structure that contains three α helices, and a sequence motif in the third helix recognizes and binds to the appropriate DNA sequence (15). Amino acid sequence comparison studies have subdivided the HD family into two classes of proteins: the typical HD class and the TALE (3-aa loop extension) class (16). KNOX proteins are members of the TALE class.
In addition to the HD, KNOX proteins contain a conserved bipartite domain in the N terminus that contains significant sequence similarity to the N-terminal domain in a family of TALE HD proteins found in animals. These proteins, collectively referred to as MEIS, include the myeloid ecotropic viral integration site (MEIS) and pre-B cell homeobox (PBX)-regulating protein 1 (PREP1) family in vertebrates and Homothorax (HTH) in Drosophila (16). This observation suggests that the MEIS and KNOX proteins were derived from a common ancestral gene, and that these domains may have similar biochemical functions. This homologous domain is referred to as the MEINOX domain (16).
The biochemical function of the MEINOX domain in MEIS proteins is to mediate heterodimerization with another group of TALE HD proteins, PBX in vertebrates and extradenticle (EXD) in Drosophila (17–21). Heterodimerization of MEIS and PBX have at least three functions in the cell. First, studies suggest that this interaction occurs in the cytoplasm, because nuclear import depends on heterodimerization (21–26). Second, like most HD proteins, MEIS and PBX monomers bind specifically to their DNA-binding motifs with low affinity; however, the affinity is greatly increased when they cooperatively bind to their appropriate DNA sequences as a heterodimer (17–21, 26). Lastly, this interaction is important for protein stability (27, 28) and in Drosophila, hth and exd mutants have strikingly similar phenotypes (22, 24, 29–31). Taken together, these studies strongly suggest that the MEIS–PBX heterodimer can be described as a single functional unit (32).
Recently two-hybrid studies have indicated that KNOX proteins interact with BEL1-like (BELL) proteins, another class of TALE HD proteins found in plants (33, 34). The mRNA expression patterns of KNOX and BELL proteins overlap in meristems, indicating that they may potentially interact in vivo. However, the biochemical significance of these interactions has not been addressed.
Our research currently is focused on the biochemical function of KN1 at the cellular level. As a first step, we characterized the biochemical function of the MEINOX domain of KN1 and found that it interacts with a TALE HD protein, knotted interacting protein (KIP). KIP is homologous to the BELL family of TALE HD proteins in Arabidopsis. The MEINOX domain of KN1 interacts with a conserved N-terminal domain in KIP. To address the biochemical function of this association, we identified the DNA-binding motifs of KN1 and KIP and demonstrate that these proteins bind with high affinity to the TGACAG(G/C)T sequence only as a complex, suggesting that the interaction of KNOX and BELL proteins is important for transcriptional activation and/or repression.
Materials and Methods
All chemicals were obtained from Sigma unless otherwise noted. All oligonucleotides were synthesized at Operon Technologies, Alameda, CA. Alignments were performed by dna star. All sequencing was performed at the University of California, Berkeley DNA Sequencing Facility. For information on plasmid construction, recombinant protein purification, and sequences of electrophoretic mobility-shift assay (EMSA) oligonucleotides see additional Materials and Methods, which are published as supporting information on the PNAS web site, www.pnas.org.
Yeast Two-Hybrid Screen.
All plasmids were transformed into the yeast strain pJ694A by using the lithium acetate method (35) then plated on the appropriate selection media. Western blot analysis using KN1 antibodies demonstrated that the GAL4-BD-MEINOX domain (where BD is the binding domain) was expressed in yeast (data not shown). The yeast two-hybrid screen was performed as described by the HYBRIDZAP 2.1 protocol from Stratagene. Plasmids were isolated from the four positive yeast colonies by using the RPM Yeast Plasmid Isolation Kit (Q-BIOgene, Carlsbad, CA), then transformed into DH5α. After transformation, plasmids were purified by using the Qiagen (Chatsworth, CA) Miniprep System and sequenced.
To identify the domain in KIP that associates with the MEINOX domain of KN1, regions of kip were cloned into pAD-GAL4 (Stratagene) and transformed into pJ694A containing the pGAL4-BD-MEINOX and plated on Trp− and Leu−. Yeast colonies containing both GAL4 plasmids were plated on His−/Ade− selection media to identify a domain(s) in KIP that interacted with the MEINOX domain in KN1.
Coimmunoprecipitation Assays.
To efficiently synthesize KIP in vitro with the TNT wheat germ extract system, we created a construct missing the first 42 aa but containing the MEINOX interacting domain (MID) and HD, *KIP43–360.
For coimmunoprecipitation assays, (i) 50 ng of the full-length purified recombinant protein was mixed with 10 μl of radiolabeled KN1 or KIP, for 2 h at 4°C in 100 μl PB buffer (20 mM Tris⋅HCl, pH 7.5/50 mM NaCl/25 mM KCl/0.2% Triton X-100/1% BSA), (ii) 100 ng of monoclonal T7 antibodies (Novagen) and 50 ng of polyclonal glutathione S-transferase (GST) antibodies were added to the binding reaction for 2 h at 4°C, (iii) 10 μl protein A and G-coated magnetic beads (Dynal, Great Neck, NY) were added to the binding reaction for 1 h at 4°C, and (iv) the beads were pelleted and washed five times in PB buffer. The bound proteins were eluted with 25 μl of SDS-loading buffer (50 mM Tris⋅HCl, pH 6.8/100 mM DTT/2% SDS/0.1% bromophenol blue/20% glycerol) then separated by 10% SDS/PAGE. The gels were dried and developed for 24 h by autoradiography using standard methods (36).
Selection and Identification of the KN1 DNA-Binding Sequence.
To synthesize a pool of degenerate double-stranded DNA (dsDNA), a single-stranded, 60-bp oligonucleotide [selection and amplification binding assay (SAAB) oligo] was synthesized (5′-GAGAGGATCCAGTCAGCATG(N)20CTCAGCCTCGAGAATTCCAA-3′) that contained 20 degenerate nucleotides flanked by 20-bp PCR primer binding sites. In addition, 3′ and 5′ primers were synthesized that were complementary to the PCR primer binding sites. BamH1 and EcoR1 restriction enzymes sites (underlined) were located on the 5′ and 3′ PCR handles, respectively. The 3′ primer was annealed to the SAAB oligo, and Klenow fragment (Panvera/Takara, Madison, WI) was used to synthesize a pool of dsDNAs. The SAAB was used to identify the KN1 DNA-binding motif as described (18). Twenty five clones identified in the screen were sequenced (see Fig. 3).
Figure 3.
Selection and amplification of the KN1 DNA-binding motif. In some cases more than one DNA motif was found on SAAB clone. * designates identical SAAB clone.
EMSA.
In EMSA, recombinant expressed and purified proteins were mixed with 50,000 cpm of wild-type DNA (WT-DNA) or mutant DNA (MT-DNA) probes, 2 μg of poly [d(I⋅C)] in 1× EMSA buffer (10 mM Tris⋅HCl, pH 7.6/75 mM NaCl/0.5 mM DTT/0.5 mM EDTA/1 mM MgCl2/5% glycerol/0.15% NP-40) for 30 min at 4°C in a 20-μl reaction volume. After binding, samples were analyzed by EMSA using 0.5× Tris-borate-EDTA polyacrylamide gels. KN1 and KN1-KIP heterodimers were analyzed on 4% polyacrylamide gels (80:1 ratio of acrylamide to bis-acrylamide), and GST-HDKIP was analyzed on 5% polyacrylamide gels (40:1 ratio of acrylamide to bis-acrylamide). After electrophoresis, the gels were dried, developed for 10–14 h, and analyzed by autoradiography.
Results
The MEINOX Domain of KN1 Interacts with a TALE HD Protein in a Yeast Two-Hybrid System.
Because MEIS and PBX depend on each other for function (32), we hypothesized that KN1 function may also depend on an interacting factor, possibly a TALE HD protein. Given that the interaction of MEIS and PBX occurs through the MEINOX domain of MEIS and a conserved domain in PBX (18, 19, 26), we sought to determine whether the MEINOX domain of KN1 functioned in a similar manner. The MEINOX domain was fused in-frame to the DNA BD of GAL4 (pGAL4-BD-MEINOX97–103) and a yeast two-hybrid library, prepared with mRNA isolated from maize inflorescence meristems where KN1 is expressed, was screened. This library was fused to the activation domain (AD) of GAL4. Of the 1 × 106 transformants screened, only four colonies grew on selective His−/Ade− media. Isolation and sequencing of these four library plasmids showed that these clones were identical. We refer to this clone as KIP.
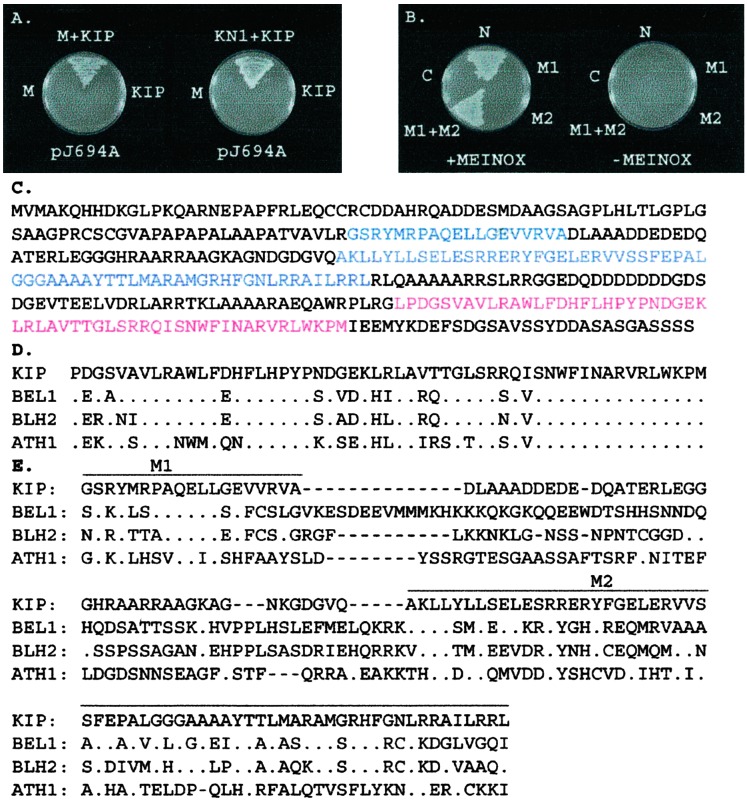
To confirm the interaction between KN1 and KIP, we retransformed the pKIP-AD-GAL4 library clone into pJ694A cells expressing pGAL4-BD-MEINOX and full-length KN1 fused to the GAL4-BD (pGAL4-BD-KN1). The results showed that KIP interacted with the MEINOX domain as well as with KN1 (Fig. 1A). In addition, pJ694A transformed with only pKIP-AD-GAL4, pGAL4-BD-MEINOX, or pGAL4-BD-KN1 did not grow on His−/Ade− selective media, indicating that these proteins do not autoactivate transcription of the selective marker genes used in our screen (Fig. 1A).
Figure 1.
Yeast two-hybrid and sequence alignments. (A) KIP interacted with the MEINOX domain (M) of KN1 and full-length KN1. (B) Interaction of the MEINOX domain with KIP depended on both conserved regions, M1 and M2, in the N terminus (N) of this protein. (C) The amino acid sequence of KIP showing M1 and M2 of the MID (blue) and HD (red). (D) Alignment of the HD sequences of KIP and BELL proteins of Arabidopsis. (E) Alignment of the MID, M1 and M2, in KIP and the Arabidopsis BELL proteins. The following GenBank accession numbers were used: BEL1, U39944; BLH2, AF173816; ATH1, X80126.
To obtain a full-length kip cDNA, we screened a maize vegetative meristem cDNA library and identified a slightly longer clone that contained four stop codons in-frame with the first methionine of the predicted protein sequence (data not shown). Sequence analysis showed that the two-hybrid cDNAs were all missing the first 9 bp of the predicted ORF (data not shown). The size of this clone is 1.4 kb, which is similar in length to the kip hybridizing transcripts determined by RNA gel blot analysis (data not shown). In addition, Southern blots showed that kip is a single copy gene that maps to chromosome 1L between restriction fragment length polymorphism markers ias7 and tbp1.
Sequence analysis showed that kip encodes a 365-aa polypeptide related to the BELL HD proteins in Arabidopsis (Fig. 1C; ref. 37). The HD of KIP contains the 3-aa extension loop, PYP motif (Fig. 1D), which is a characteristic feature of TALE HD proteins (16). Sequence alignment by clustal x of KIP and the Arabidopsis BELL proteins showed KIP to be 72–85% identical to these proteins in the HD (Fig. 1D).
The MEINOX Domain of KN1 Interacts with a Conserved Region in the N Terminus of KIP.
To determine the region(s) in KIP that were necessary for interaction with the MEINOX domain of KN1, we tested whether the N-terminal or C-terminal halves of KIP could interact with the MEINOX domain in the yeast two-hybrid system. Results show that only the N-terminal region, which does not contain the HD, interacted with the MEINOX domain (Fig. 1B). Sequence alignment of the N termini of KIP and the Arabidopsis BELL proteins revealed two regions of conservation (Fig. 1E; M1 and M2). We tested whether or not M1 or M2 could interact with the MEINOX domain and found that neither region alone was sufficient (Fig. 1B). However, when we tested the region that contained both M1 and M2 we found that this domain was sufficient and necessary for interaction with the MEINOX domain (Fig. 1B). Conservation of M1 and M2 in BELL proteins (Fig. 1E) strongly suggests that the function of this domain is to interact with the MEINOX domain in KNOX proteins. In fact, the M1 and M2 regions in BEL1, also known as SKY and BELL domains, respectively, were shown to interact with KNAT1 and KNAT3 (33). Therefore, we refer to this region as MID.
KN1 and KIP Interact in Vitro.
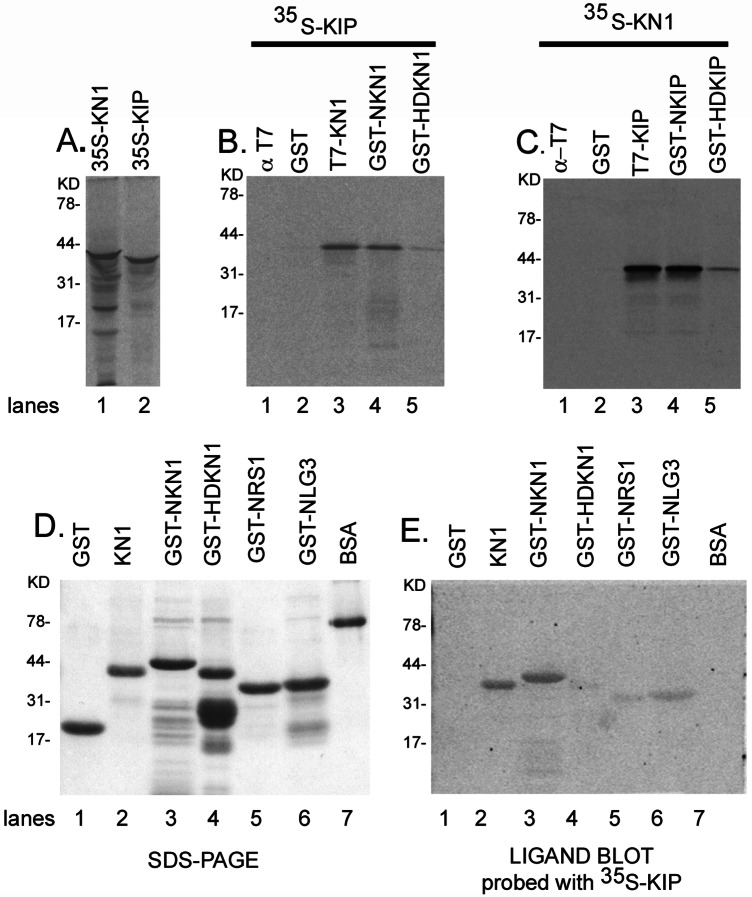
To confirm our yeast two-hybrid studies, we performed an in vitro binding/coimmunoprecipitation assay, using recombinantly expressed and purified KN1 and KIP proteins. Protein fusion tags were used in our experiments to not only affinity-purify KN1 and KIP, but to immunoprecipitate them as well. Full-length KN1 (T7-KN1) and KIP (T7-KIP) contained an N-terminal T7 epitope tag and a C-terminal 6× histidine tag. These proteins were purified from soluble bacterial extracts by Ni chromatography. The N- and C-terminal regions of KN1 and KIP were cloned in-frame with GST for glutathione affinity chromatography. The N-terminal GST fusions of KN1 (GST-NKN1) and KIP (GST-NKIP) contained the conserved MEINOX domain and MID, respectively. The C-terminal GST fusions of KN1 (GST-HDKN1) and KIP (GST-HDKIP) contained the HDs.
Fig. 2A shows the in vitro-translated 35S-methionine (Met)-labeled KN1 and *KIP43–360 proteins that were separated by SDS/PAGE and analyzed by autoradiography. In our binding reaction, radiolabeled KN1 and KIP were mixed with purified KIP and KN1 fusion proteins, respectively. T7 antibodies were used to immunoprecipitate full-length KN1 and KIP, and GST antibodies were used to immunoprecipitate the GST N- and C-terminal fusion proteins of KN1 and KIP. In Fig. 2B, [35S]Met-*KIP43–360 coimmunoprecipitated with T7-KN1 and GST-NKN1, whereas minimal interaction was detected with GST-HDKN1. Coimmunoprecipitation results also demonstrate that [35S]Met-KN1 interacted with full-length KIP and N-GSTKIP whereas less association was detected with GST-HDKIP (Fig. 2C). In control experiments, neither radiolabeled protein interacted with GST or could be immunoprecipitated with T7 (Fig. 2 B and C). Also, GST antibodies could not immunoprecipitate the radiolabeled proteins (data not shown). These in vitro binding results demonstrate that the N-terminal regions of KN1 and KIP are sufficient for interaction and support our results from the yeast two-hybrid experiments (Fig. 1).
Figure 2.
In vitro binding studies with KIP and KNOX proteins. (A) In vitro-synthesized [35S]Met-KN1 and KIP were analyzed by SDS/PAGE. (B and C) Coimmunoprecipitation was used to analyze the interaction of KIP and KN1. The ligand blot assay used in our experiments has been described (49, 50). About 1 μg of each protein fusion as well as GST and BSA controls were separated by SDS/PAGE (D) and blotted to nitrocellulose. After the proteins were denatured and renatured, the blots were probed with 20 μl of [35S]Met-KIP in renaturation buffer for 4 h at 4°C, gently shaking. Blots were washed in Tris buffer solution plus 0.05% Tween 20 (49), air-dried, developed for 12 h, and analyzed by autoradiography (E).
KIP Has a Higher Affinity for KN1 than RS1 and LG3.
Given that other knox genes, such as rs1 and lg3, are expressed in maize meristems (2), we asked whether KIP could also interact with these proteins. To address this question, we used a ligand blot assay in which recombinant purified fusion proteins were separated by SDS/PAGE, blotted to nitrocellulose, and probed with [35S]Met-*KIP43–360. GST N-terminal fusions that contained the MEINOX domains of KN1 (GST-NKN1), RS1 (GST-NRS1), and LG3 (GST-NLG3) were prepared (Fig. 2 D and E). In addition, we tested whether or not labeled KIP could interact with “full-length” KN1 and GST-HDKN1. Ligand blot analysis demonstrated that [35S]Met-*KIP43–360 bound to KN1 and GST-NKN1, whereas little or no interaction was detected with GST-HD, GST, and BSA (Fig. 2E). Interestingly, there was little association of [35S]Met-*KIP43–360 with GST-NRS1 or GST-NLG3 (Fig. 2E), suggesting that KIP has an apparent higher affinity for KN1 than the other KNOX proteins.
Identification of the KN1 DNA-Binding Motif.
Because KN1 and KIP encode HDs, the expectation exists that they bind to specific DNA sequences. We therefore sought to identify the DNA-binding motifs for KN1 and KIP to determine how the interaction of KN1 and KIP affected DNA binding.
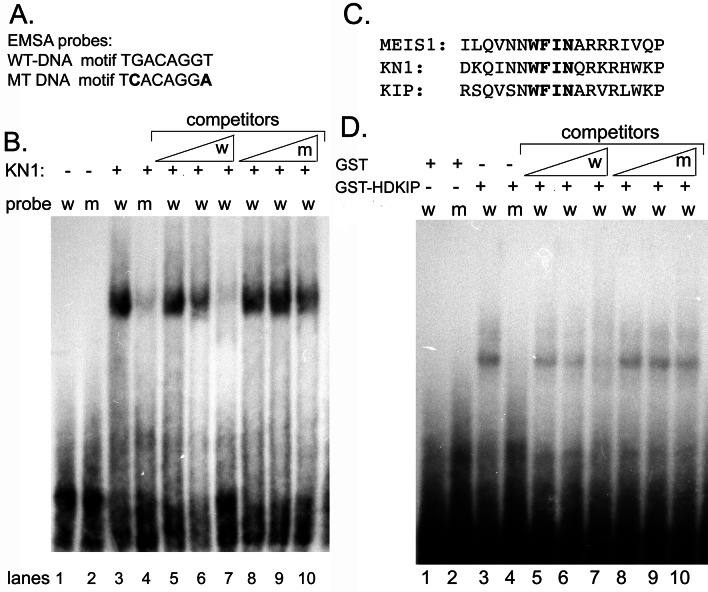
The SAAB was used to biochemically identify the DNA-binding motif for KN1 (18, 38). A pool of dsDNA molecules was synthesized, containing two 20-nt PCR primer binding sites at the 5′ and 3′ ends of each molecule separated by 20 degenerate nucleotides. Purified T7-KN1 was incubated with the dsDNA, and the DNA–protein complexes were immunoprecipitated with T7 mAbs. With the 5′ and 3′ primers, the eluted DNA molecules were amplified by PCR. The amplified DNA molecules were then mixed with purified T7-KN1 for another round of selection and amplification. This cycle was repeated six times to enrich for KN1 DNA-binding sites, and the final amplified DNA products were cloned and sequenced. In a control experiment in which T7 antibodies were mixed with the dsDNA, we were unable to immunoprecipitate and amplify any DNA, demonstrating that the T7 antibodies do not bind to DNA (data not shown). The DNA-binding motif identified for KN1 was identical to the MEIS binding site, TGACAG(G/C)T (Fig. 3; ref. 18).
KN1 Binds Specifically to the TGACAG(G/C)T Sequence.
To verify the SAAB results from above, EMSAs were used to investigate the DNA-binding ability and specificity of KN1. Complementary oligonucleotides were synthesized for clone 15 (WT-DNA; Fig. 4), which contained the TGACAGGT binding motif. Studies in animals showed that the second nucleotide, G, in the MEIS binding motif is critical for DNA association and changing it to cytosine reduced most of the MEIS–DNA interaction (18). Therefore, to determine specificity, we synthesized complementary oligonucleotides that contained a cytosine at position 2 and an adenine at position 8 (MT-DNA) in clone 15 so that the mutated binding motif had the following sequence, TCACAGGA (Fig. 4A). To reconstitute DNA binding in EMSA, we used high concentrations (at least 100 ng) of T7-KN1 protein. At these concentrations, T7-KN1 bound to the WT-DNA probe, whereas little DNA-binding activity was detected with the MT-DNA probe (Fig. 4B, lane 3 and 4).
Figure 4.
EMSA analysis of purified T7-KN1 and GST-HDKIP. (A) WT-DNA and MT-DNA binding sites were used as probes. (B) T7-KN1 was incubated with WT-DNA probe (W) or MT-DNA probe (M). (C) The third helices in the HDs of MEIS, KN1, and KIP were aligned. (D) In EMSA, GST or GST-HDKIP was incubated with W or M probes. (B and D) DNA-binding specificity for T7-KN1 and GST-HDKIP were determined by competing with unlabeled WT-DNA (W) or MT-DNA (M), at 5-, 50-, and 500-fold molar excess over labeled probe.
To verify the specificity of the DNA binding, increasing amounts of unlabeled WT-DNA or MT-DNA were added to the binding reaction. Our results show that unlabeled WT-DNA competed for DNA binding, whereas the MT-DNA competitor had little effect (Fig. 4B, lanes 5–10). When other clones such as 5, 10, and 14 were used as probes in EMSA, we found that T7-KN1 specifically bound to these probes with equal intensities (data not shown), showing that the nucleotides surrounding the TGACAG(G/C)T binding sequence are not important for binding of the T7-KN1 protein. Lastly, recombinant KN1, which does not contain a T7 or His tag, specifically associated with the DNA-binding motif (data not shown), demonstrating that these tags are not involved in DNA binding. The addition of KN1 antibodies to the binding reaction resulted in supershifted DNA–protein complex in EMSA whereas control sera had little effect on the mobility of the KN1–DNA complex (data not shown). Therefore, we conclude that KN1 specifically binds to the TGACAG(G/C)T sequence.
The HD of KIP Specifically Recognizes the KN1 DNA-Binding Motif.
The third helix of HD proteins contains the conserved WF_N sequence. Based on biochemical and structural studies, variability in the amino acid that occupies the third position in the WF_N sequence is important for determining the DNA-binding sequence specificity (39). MEIS, KN1, and KIP all contain the WFIN sequence in the third helix (Fig. 4C), and because MEIS and KN1 specifically recognize the TGACAG(G/C)T sequence, KIP may associate with this motif as well. We investigated this possibility by using EMSA, but we could not reconstitute DNA binding with a full-length T7-KIP, because this protein was insoluble and expressed at low levels in bacteria (data not shown). Although we were able to purify small amounts of T7-KIP, we did not detect DNA binding at low concentrations (Fig. 5A, lanes 2 and 5). Studies in animals have demonstrated that the HD alone binds to its appropriate DNA-binding motif (15). Therefore, we used the GST-HDKIP to address whether or not KIP can associate with the KN1 DNA-binding motif.
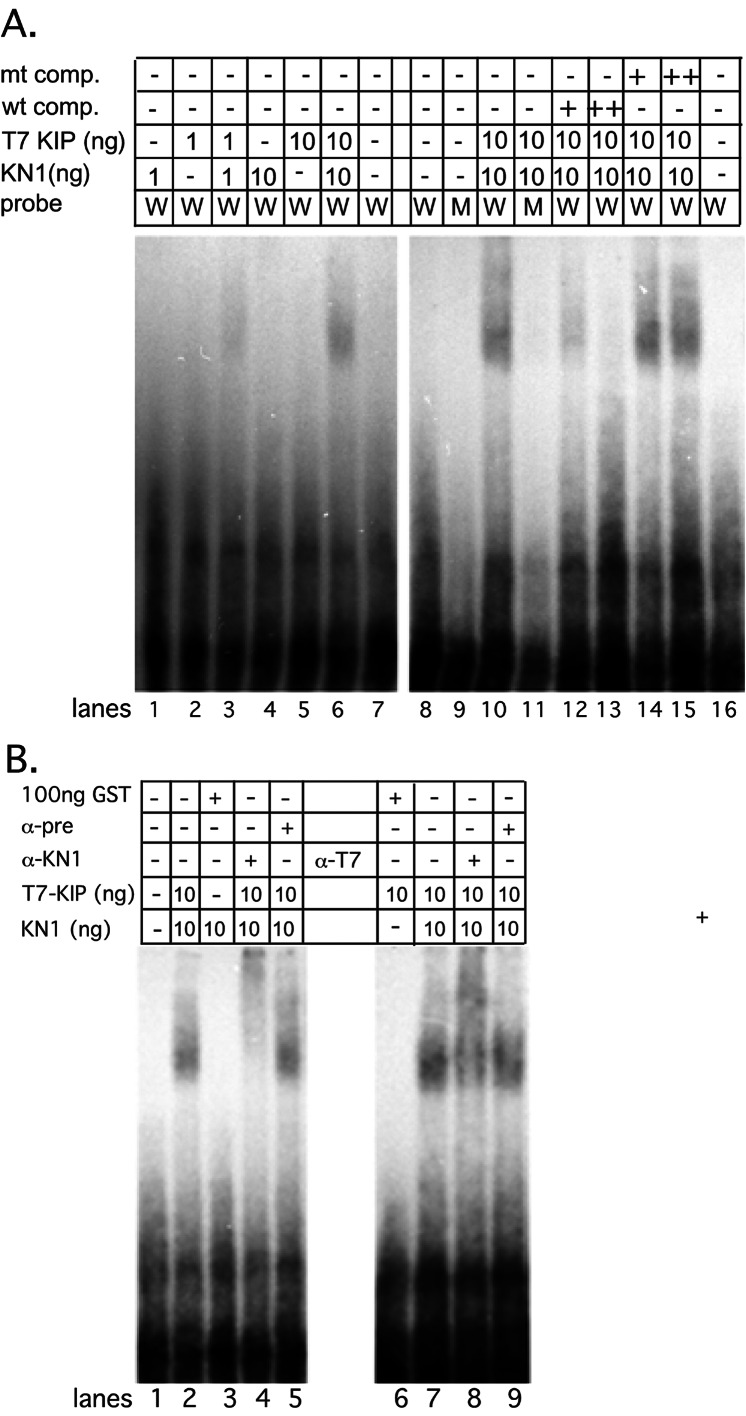
Figure 5.
EMSA analysis of the KN1–KIP complex. (A) DNA-binding assays were performed with 1 ng or 10 ng of KN1 or T7-KIP alone. KN1 and T7-KIP were mixed together in DNA binding assays at 1 ng or 10 ng of each protein per reaction. DNA-binding specificity was determined by competing with unlabeled WT-DNA (wt comp) or MT-DNA (mt comp), at 10- and 100-fold molar excess over labeled probe. (B) Antibodies to KN1 (α-KN1) or the T7 epitope (α-T7) were added to the DNA-binding reaction. As a control, preimmune (α-pre) sera were also added to the KN1-KIP DNA-binding reaction. Mixing 100 ng GST with 10 ng KN1 or 10 ng of KIP did not form a DNA-binding complex.
Reconstitution of DNA binding with 200 ng of GST-HDKIP demonstrated that the HD of KIP bound to the WT-DNA probe, whereas little association was detected with the MT-DNA (Fig. 4D, lane 3 and 4). Fig. 4D shows that GST doesn't bind to either DNA probe. In competition experiments, unlabeled WT-DNA competed for DNA binding, whereas unlabeled MT-DNA had little effect (Fig. 4D, lanes 5–10). The addition of GST antibodies to the binding reaction reduced the mobility of the GST–HDKIP–DNA complex in EMSA, whereas control sera had no effect on the mobility of this complex (data not shown). These results indicate that the HD of KIP specifically interacts with the KN1-binding motif.
The KN1–KIP Complex Binds to the TGACAGGT Sequence with a Higher Affinity than KN1 and KIP Alone.
To determine how the interaction of KN1 and KIP could affect DNA binding to the TGACAGGT sequence, we performed DNA-binding experiments combining KN1 and KIP proteins. Fig. 5A shows that at low concentrations of full-length KN1 (lanes 1 and 4) or T7-KIP (lanes 2 and 5), we were unable to reconstitute DNA binding. However, when low concentrations of KN1 and T7-KIP were incubated together with WT-DNA probe, we detected association (Fig. 5A, lanes 3, 6, and 10). No association was detected with MT-DNA probe (Fig. 5A, lane 11). In competition experiments, only the unlabeled WT-DNA reduced the binding, whereas MT-DNA had little effect (Fig. 5A, lanes 12–15). Interestingly, lower concentrations of competitor could compete for binding with the KN1–KIP complex than with KN1 or GST-HDKIP alone.
To confirm that KN1 and KIP were cooperatively binding to the TGACAGGT motif, antibodies directed against KN1 or the T7 epitope tag (found only on KIP) were added to the DNA-binding reaction. In EMSA, KN1 and T7 antibodies reduced the mobility of the DNA–protein complex (Fig. 5B, lanes 4 and 8), whereas control sera had no effect on the mobility of the DNA–protein complex (Fig. 5B, lanes 5 and 9). In addition, when low concentrations of KN1 or KIP were mixed with GST, no DNA–protein interaction was detected by EMSA (Fig. 5B, lanes 3 and 6). Taken together, these results demonstrate that a KN1–KIP complex binds specifically to the TGACAGGT sequence with an apparent high affinity.
Discussion
We have found that KN1 interacts with KIP, a member of the BELL family of plant TALE HD proteins. This association is mediated by a conserved MEINOX domain in KN1 and MID in KIP. Because the MEINOX domain is required for interactions among TALE HDs in animals and plants, the results demonstrate that the biochemical function of the MEINOX domain is evolutionarily conserved. In vitro binding studies suggest that KIP may selectively bind to KN1 rather than to other KNOX proteins in the SAM. This observation is also supported by the fact that [35S]Met-KIP selectively binds to STM, the KN1 ortholog, with a higher affinity than to KNAT1 or KNAT2 in Arabidopsis ligand blot assays (unpublished data). In Arabidopsis, BEL1 may also selectively bind to a subset of KNOX proteins (33). Taken together, these studies strongly suggest that the interaction of KNOX and BELL proteins depends on not only overlapping expression patterns but also on the ability of these proteins to physically associate with one another.
Although KNOX–BELL interactions have been described in Arabidopsis (33) and barley (34), the biochemical function of this interaction was not addressed. Because DNA recognition and binding are initial steps in gene regulation, we examined how the interaction of KN1 and KIP affects DNA binding. We explored this question by using in vitro DNA-binding assays. As a first step, we identified the KN1 DNA-binding motif and found that both KN1 and the HD of KIP bound specifically to this motif with low affinity. Moreover, we showed that cooperative interaction of KN1 and KIP dramatically increased the DNA-binding affinity, suggesting that selective interaction of KNOX and BELL proteins could be important for gene regulation. Until KN1–KIP target genes are identified and thoroughly characterized, we cannot, at the present moment, address in vivo function of this TALE complex. Nevertheless, analysis of the KN1–KIP complex in vitro is essential toward functional studies in vivo.
In animals and yeast, TALE HD proteins form complexes that cooperatively bind to their appropriate DNA-binding motifs with high affinity, whereas the monomeric forms bind with low affinity (17–21, 26, 40). Moreover, these in vitro DNA-binding observations correlate with in vivo function (41, 42). Combinatorial interaction of TALE HD proteins appears to not only be evolutionarily conserved but might be necessary for regulatory control of developmental pathways. Therefore, because the interaction of KN1 and KIP dramatically increases the DNA-binding affinity similar to other heterodimeric TALE–TALE HD interactions, it suggests that association of KNOX and BELL proteins might be important steps in controlling developmental pathways that are crucial for SAM function.
Analysis of the third helix of the HD shows that amino acid residues, WFIN, involved in DNA recognition, are conserved in all MEINOX containing proteins in plants and animals, as well as the BELL family in plants. These results suggest that all KNOX and BELL proteins bind to the same DNA-binding motif. Previous studies have identified DNA-binding sites for other KNOX proteins in barley (43), tobacco (44), and rice (45) that share some similarity with the KN1 binding site. In fact, the TGAC half site is absolutely conserved in all of the above studies. This half site is also critical for DNA binding of the MEIS protein. Slight variations in these TALE HD DNA-binding motifs suggest that the 3′ end of the binding site has less effect on binding than the 5′ end of the sequence. Because the WFIN sequence is conserved in MEIS and KNOX proteins, and the TGACAG(G/C)T sequence was selected for MEIS1 (18) and KN1 (Fig. 3), we suggest that this DNA sequence is more efficiently recognized by these classes of TALE HD proteins.
Genetic and molecular studies have shown that KNOX proteins function in the SAM; however, the function of BELL genes in the SAM is unclear. The bel1 mutant in Arabidopsis is the only characterized mutant from this gene family, and its defect suggests that it functions in ovule development (37). Interestingly, although bel1 mutants produce a similar number of internodes as WT, the inflorescence meristem will sometimes terminate with carpelloid structures (46). This observation suggests that BEL1 may function in the SAM, and recent studies have shown that BEL1 is expressed at low levels in this tissue as well (33).
Expression analysis suggests that KN1 and KIP could interact in the SAM (Fig. 6, which is published as supporting information on the PNAS web site). Interestingly, kip is also expressed in young leaves where kn1 is not expressed (Fig. 6). It is also of interest that BEL1 is expressed in leaves, too, yet there is no obvious effect in bel1 mutant leaves (37).
As a model, we propose that KNOX–BELL interactions form a basic transcriptional unit that is important for specifying cell fate in the meristem. Our results strongly indicate that all KNOX and BELL proteins recognize the same DNA-binding motif; however, genetic and molecular evidence in Arabidopsis suggests that STM and KNAT1 control different developmental pathways (3, 4, 47). Even if there is functional overlap between these two knox genes, it is difficult to explain how they control different pathways (i.e., target genes) yet bind to the same DNA sequence. In animals, MEIS–PBX heterodimers can also serve as cofactors for a subset of typical HD proteins (HOX) (32). HOX proteins bind to DNA with low affinity and specificity as monomers; however, a trimeric complex of MEIS–PBX–HOX is essential for in vivo function and high affinity DNA binding (26, 41, 42, 48). Therefore, we speculate that KNOX–BELL complexes could function as cofactors for plant HOX proteins to control cell fate.
Supplementary Material
Acknowledgments
We thank Dr. David Barnes (University of Wisconsin, Madison) for the pJ69-4A yeast strain, Dr. Michael Freeling (University of California, Berkeley) for the rs1 and lg3 cDNAs, and Dr. Robert Schmidt (University of California, San Diego) for the immature ear yeast two-hybrid library. Thanks to George Chuck, Enamul Huq, and Peter Quail for critical reading of this manuscript. H.M.S.S. was supported by National Institutes of Health Postdoctoral Fellowship GM20158-03. S.H. was supported by National Science Foundation Grant MCB-9727611 and the U.S. Department of Agriculture.
Abbreviations
- kn1
knotted1
- rs1
rough sheath1
- lg3
liguleless3
- KIP
knotted interacting protein
- STM
SHOOTMERISTEMLESS
- MEIS
myeloid ecotropic viral integration site
- PBX
pre-B cell homeobox
- TALE
3-aa acid loop extension
- SAM
shoot apical meristem
- EMSA
electrophoretic mobility-shift assay
- BELL
BEL-like protein
- MID
MEINOX interacting domain
- GST
glutathione S-transferase
- WT-DNA
wild-type DNA
- MT-DNA
mutant DNA
- BD
binding domain
- HD
homeodomain
- AD
activation domain
- SAAB
selection and amplification binding assay
- dsDNA
double-stranded DNA
Footnotes
Data deposition: The sequence of kip reported in this paper has been deposited in the GenBank database (accession no. AY082396).
References
- 1.Reiser L, Sanchez-Baracaldo P, Hake S. Plant Mol Biol. 2000;42:151–166. [PubMed] [Google Scholar]
- 2.Jackson D, Veit B, Hake S. Development (Cambridge, UK) 1994;120:405–413. [Google Scholar]
- 3.Long J A, Moan E I, Medford J I, Barton M K. Nature (London) 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- 4.Barton M K, Poethig R S. Development (Cambridge, UK) 1993;119:823–831. [Google Scholar]
- 5.Vollbrecht E, Reiser L, Hake S. Development (Cambridge, UK) 2000;127:3161–3172. doi: 10.1242/dev.127.14.3161. [DOI] [PubMed] [Google Scholar]
- 6.Endrizzi K, Moussian B, Haecker A, Levin J Z, Laux T. Plant J. 1996;10:101–113. doi: 10.1046/j.1365-313x.1996.10060967.x. [DOI] [PubMed] [Google Scholar]
- 7.Clark S E, Jacobsen S E, Levin J Z, Meyerowitz E M. Development (Cambridge, UK) 1996;122:1567–1575. doi: 10.1242/dev.122.5.1567. [DOI] [PubMed] [Google Scholar]
- 8.Kerstetter R A, Laudencia-Chingcuanco D, Smith L G, Hake S. Development (Cambridge, UK) 1997;124:3045–3054. doi: 10.1242/dev.124.16.3045. [DOI] [PubMed] [Google Scholar]
- 9.Smith L G, Greene B, Veit B, Hake S. Development (Cambridge, UK) 1992;116:21–30. doi: 10.1242/dev.116.1.21. [DOI] [PubMed] [Google Scholar]
- 10.Schneeberger R G, Becraft P W, Hake S, Freeling M. Genes Dev. 1995;9:2292–2304. doi: 10.1101/gad.9.18.2292. [DOI] [PubMed] [Google Scholar]
- 11.Muehlbauer G J, Fowler J E, Girard L, Tyers R, Harper L, Freeling M. Plant Physiol. 1999;119:651–662. doi: 10.1104/pp.119.2.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foster T, Yamaguchi J, Wong C B, Veit B, Hake S. Plant Cell. 1999;11:1239–1252. doi: 10.1105/tpc.11.7.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freeling M. Dev Biol. 1992;153:44–58. doi: 10.1016/0012-1606(92)90090-4. [DOI] [PubMed] [Google Scholar]
- 14.Vollbrecht E, Veit B, Sinha N, Hake S. Nature (London) 1991;350:241–243. doi: 10.1038/350241a0. [DOI] [PubMed] [Google Scholar]
- 15.Gehring W J, Affolter M, Burglin T. Annu Rev Biochem. 1994;63:487–526. doi: 10.1146/annurev.bi.63.070194.002415. [DOI] [PubMed] [Google Scholar]
- 16.Burglin T R. Nucleic Acids Res. 1997;25:4173–4180. doi: 10.1093/nar/25.21.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bischof L J, Kagawa N, Moskow J J, Takahashi Y, Iwamatsu A, Buchberg A M, Waterman M R. J Biol Chem. 1998;273:7941–7948. doi: 10.1074/jbc.273.14.7941. [DOI] [PubMed] [Google Scholar]
- 18.Chang C-P, Jacobs Y, Nakamura T, Jenkins N A, Copeland N G, Cleary M L. Mol Cell Biol. 1997;17:5679–5687. doi: 10.1128/mcb.17.10.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knoepfler P S, Calvo K R, Chen H, Antonarakis S E, Kamps M P. Proc Natl Acad Sci USA. 1997;94:14553–14558. doi: 10.1073/pnas.94.26.14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berthelsen J, Zappavigna V, Mavilio F, Blasi F. EMBO J. 1998;17:1423–1433. doi: 10.1093/emboj/17.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berthelsen J, Kilstrup-Nielsen C, Blasi F, Mavilio F, Zappavigna V. Genes Dev. 1999;13:946–953. doi: 10.1101/gad.13.8.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rieckhof G, Casares F, Ryoo H D, Abu-Shaar M, Mann R S. Cell. 1997;91:171–183. doi: 10.1016/s0092-8674(00)80400-6. [DOI] [PubMed] [Google Scholar]
- 23.Jaw T J, You L R, Knoepfler P S, Yao L C, Pai C Y, Tang C Y, Chang L P, Berthelsen J, Blasi F, Kamps M P, Sun Y H. Mech Dev. 2000;91:279–291. doi: 10.1016/s0925-4773(99)00316-0. [DOI] [PubMed] [Google Scholar]
- 24.Pai C Y, Kuo T S, Jaw T J, Kurant E, Chen C T, Bessarab D A, Salzberg A, Sun Y H. Genes Dev. 1998;12:435–446. doi: 10.1101/gad.12.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abu-Shaar M, Ryoo H D, Mann R S. Genes Dev. 1999;13:935–945. doi: 10.1101/gad.13.8.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ryoo H D, Marty T, Casares F, Affolter M, Mann R S. Development (Cambridge, UK) 1999;126:5137–5148. doi: 10.1242/dev.126.22.5137. [DOI] [PubMed] [Google Scholar]
- 27.Abu-Shaar M, Mann R S. Development (Cambridge, UK) 1998;125:3821–3830. doi: 10.1242/dev.125.19.3821. [DOI] [PubMed] [Google Scholar]
- 28.Waskiewicz A J, Rikhof H A, Hernandez R E, Moens C B. Development (Cambridge, UK) 2001;128:4139–4151. doi: 10.1242/dev.128.21.4139. [DOI] [PubMed] [Google Scholar]
- 29.Peifer M, Wieschaus E. Genes Dev. 1990;4:1209–1223. doi: 10.1101/gad.4.7.1209. [DOI] [PubMed] [Google Scholar]
- 30.Rauskolb C, Peifer M, Weischaus E. Cell. 1993;74:1101–1112. doi: 10.1016/0092-8674(93)90731-5. [DOI] [PubMed] [Google Scholar]
- 31.Kurant E, Pai C Y, Sharf R, Halachmi N, Sun Y H, Salzberg A. Development (Cambridge, UK) 1998;125:1037–1048. doi: 10.1242/dev.125.6.1037. [DOI] [PubMed] [Google Scholar]
- 32.Mann R S, Morata G. Annu Rev Cell Dev Biol. 2000;16:243–271. doi: 10.1146/annurev.cellbio.16.1.243. [DOI] [PubMed] [Google Scholar]
- 33.Bellaoui M, Pidkowich M S, Samach A, Kushalappa K, Kohalmi S E, Modrusan Z, Crosby W L, Haughn G W. Plant Cell. 2001;13:2455–2470. doi: 10.1105/tpc.010161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muller J, Wang Y, Franzen R, Santi L, Salamini F, Rohde W. Plant J. 2001;27:13–23. doi: 10.1046/j.1365-313x.2001.01064.x. [DOI] [PubMed] [Google Scholar]
- 35.Gietz R D, Schiestl R H. Yeast. 1991;7:253–263. doi: 10.1002/yea.320070307. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Press; 1989. [Google Scholar]
- 37.Reiser L, Modrusan Z, Margossian L, Samach A, Ohad N, Haughn G W, Fischer R L. Cell. 1995;83:735–742. doi: 10.1016/0092-8674(95)90186-8. [DOI] [PubMed] [Google Scholar]
- 38.Blackwell T K, Weintraub H. Science. 1990;250:1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- 39.Treisman J, Gonczy P, Vashishtha M, Harris E, Desplan C. Cell. 1989;59:553–562. doi: 10.1016/0092-8674(89)90038-x. [DOI] [PubMed] [Google Scholar]
- 40.Herskowitz I. Nature (London) 1989;342:749–757. doi: 10.1038/342749a0. [DOI] [PubMed] [Google Scholar]
- 41.Ferretti E, Marshall H, Popperl H, Maconochie M, Krumlauf R, Blasi F. Development (Cambridge, UK) 2000;127:155–166. doi: 10.1242/dev.127.1.155. [DOI] [PubMed] [Google Scholar]
- 42.Jacobs Y, Schnabel C A, Cleary M L. Mol Cell Biol. 1999;19:5134–5142. doi: 10.1128/mcb.19.7.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krusell L, Rasmussen I, Gausing K. FEBS Lett. 1997;408:25–29. doi: 10.1016/s0014-5793(97)00382-7. [DOI] [PubMed] [Google Scholar]
- 44.Sakamoto T, Kamiya N, Ueguehi-Tanaka M, Iwahori S, Matsuoka M. Genes Dev. 2001;15:581–590. doi: 10.1101/gad.867901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagasaki H, Sakamoto T, Sato Y, Matsuoka M. Plant Cell. 2001;13:2085–2098. doi: 10.1105/TPC.010113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Modrusan Z, Reiser L, Feldmann K A, Fischer R L, Haughn G W. Plant Cell. 1994;6:333–349. doi: 10.1105/tpc.6.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Douglas S J, Chuck G, Dengler R E, Pelecanda L, Riggs C D. Plant Cell. 2002;14:1–13. doi: 10.1105/tpc.010391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berthelsen J, Zappavigna V, Ferretti E, Mavilio F, Blasi F. EMBO J. 1998;17:1434–1445. doi: 10.1093/emboj/17.5.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen M H, Sheng J, Hind G, Handa A K, Citovsky V. EMBO J. 2000;19:913–920. doi: 10.1093/emboj/19.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Radu A, Blobel G, Moore M S. Proc Natl Acad Sci USA. 1995;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.