Abstract
Why do closely related primate genera vary in longevity, and what does this teach us about human aging? Life tables of female baboons (Papio hamadryas) in two wild populations of East Africa and in a large captive population in San Antonio, Texas, provide striking similarities and contrasts to human mortality patterns. For captive baboons at the Southwest Foundation for Biomedical Research, we estimate the doubling time of adult mortality rate as 4.8 years. Wild females in free-living populations in Tanzania and in Kenya showed doubling times of 3.5 and 3.8 years, respectively. Although these values are considerably faster than the estimates of 7–8 years for humans, these primates share a demographic feature of human aging: within each taxon populations primarily vary in the level of Gompertz mortality intercept (frailty) and vary little in the demographic rate of aging. Environmental and genetic factors within taxa appear to affect the level of frailty underlying senescence. In contrast, primate taxa are differentiated by rates of demographic aging, even if they cannot be characterized by species-specific lifespan.
Modern human life expectancy now extends into the 7th decade or beyond. This longevity arises from reduced mortality throughout life, including among the oldest old (1). These well documented changes raise questions central to the evolutionary theory of senescence and to public policy. First, how does human life expectancy relate to the lifespan of other primate genera, and in general how has human longevity evolved? Second, how long will humans live in future decades and how healthy will they be? At the same time, the genetic analysis of human longevity is rapidly advancing. Human adult longevity is a heritable trait where genetic factors can be mapped to defined chromosomal regions (2–4). What will we learn about aging when the genes underlying variation in lifespan are identified?
Although modern records of adult human lifespan and mortality are clear, the notion of what constitutes our natural or ancestral longevity is ambiguous. Evidence from osteological analysis of mortuary samples suggests that ancestral human adults lived until age 36 years on average and rarely beyond age 50 (refs. 5 and 6, but see ref. 7). Studies of precontact hunter–gatherers suggest that humans naturally possess greater longevity. On average, 15-year-olds of the Dobe !Kung survive to 69 years of age (8). Forest dwelling Ache at 15 live to 52 (males) and 58 (females) years (9). As recently as 1900 in the U.S., a 15-year-old could expect to live to 62 years; this value has gradually extended to its current state of 77 years (combined for males and females and all races; ref. 10). The variability among these observations illustrates a fundamental challenge if we are to understand the evolution of human senescence; we must fix a reference point to describe how humans and other primates age in the absence of modern environment and cultural practice.
Comparative analyses of senescence have addressed this variability with different approaches. Recently, Judge and Carey compiled data on observed record lifespans of 53 primate genera (11). Variance among genera in record lifespan was explained largely by brain mass and body size, and with this model Homo maximum longevity was predicted to occur between 52 and 90 years. They concluded that human longevity reflects a phylogenetic legacy associated with the evolution of body size and encephalization.
Finch et al. (12, 13) illustrated an alternative comparative approach. As an empirical observation, mortality rate μx increases as an exponential function of adult age where μx = λeγx. Although the rate of change in μx can decelerate among the oldest old in many species, deceleration is not described for humans less than 80 years of age (14). The parameter γ regulates the rate of change in mortality with age and is referred to as the demographic rate of aging. In contrast, the parameter λ, the baseline or initial adult mortality rate, enjoys less clarity. Vaupel et al. (15) described λ as the mean fixed individual frailty initially present in a cohort; frailty itself is the set of susceptibilities and risk factors that alter an individual's chance of death at specific ages. Sacher (16) argued that λ represents individual vulnerability to disease or physiological damage. Concerning γ, Finch et al. (12) estimated the mortality rate doubling time (MRDT = ln2/γ) and found relative constancy among groups of humans, 7–8.5 years. Based on three contemporary female life tables, the estimate of γ by Gage (17) all produce MRDT between 6 and 7 years. In contrast, in all cases the baseline mortality varied markedly among populations. Under the provisional assumption that γ is resistant to environmental conditions, Finch et al. (12) concluded that the mortality rate accelerations estimated for other mammals are similar to the slow rate observed for humans.
Extensive life tables of the baboon Papio hamadryas both in the wild (East Africa) and in captivity (Texas) offer the first opportunity to provide replicated, independent estimates of mortality rates for a non-human primate species. In Gombe National Park, Tanzania, females have been observed since 1968. As previously reported, demographic aging is evident in the Gombe population because adult mortality accelerates with age (18). Here we update the Gombe life table to reduce the level of censoring and we estimate the Gompertz mortality parameters of demographic aging. In Kenya, baboons resident in Amboseli National Park and environs have been followed continuously since 1971, and extensive demographic data are available on the population (19, 20). Here we analyze adult mortality rates and estimate Gompertz parameters of the Amboseli females. At Southwest Foundation for Biomedical Research (SFBR), Texas, records of birth and death with natural cause have been compiled from more than 8,000 females and 6,000 males since 1964. From these data (including extant cohorts), we develop life tables for both males and females and estimate parameters of mortality trajectory. We use these life tables to make the first comparison of mortality trajectory among multiple populations of a non-human primate, compare aging in wild and captive animals, and provide a reference point for contrasts of demographic aging to Homo.
Methods
Study Populations.
Population of SFBR.
Baboons of SFBR are Papio hamadryas anubis (olive), P. hamadryas cynocephalus (yellow), and their hybrids. The colony was established in 1964 with wild-caught animals from Kenya and Tanzania and is maintained at a stable size of about 3,000 individuals. Animals represented in this life table were housed in a collection of social cages each with about 20 individuals comprised of multiple adult females, a single adult male, and numerous juveniles less than 6 months old (practice before 1999; current removal age is 9 months). Weaned juveniles were reared in same-age groups indoors during the winter and outdoors during the summer; proximal to age of maturation, individuals were placed in social cages with adults. The social cages were outdoors with sheltered areas and measure 500 or 1,000 square feet. Animals were fed commercial monkey chow ad libitum and water, checked twice daily for overall health and status (disease, injury, pregnancy/menstrual cycle status), and examined with extensive physicals once or twice a year. Animal maintenance and care conformed to the ethical guidelines of the Institutional Animal Care and Use Committee.
Female vital statistics of SFBR are compared with female data from the wild populations; statistics on males are restricted to the SFBR males. Birth, death, acquisition, removal, and vital data are maintained in a colony database. Of 5,814 colony-born females, 2,113 individuals were excluded because of their participation in medical trials or their release from SFBR. Of the 3,801 included females, 1,670 had died while resident in the colony, and 2,131 were alive at our census date (30 October 2000) and thus were right censored. Similarly, 4,247 of 6,806 males were excluded, 1,303 died, and 1,256 were right censored. Deaths included in the study were only those from natural causes or euthanasia when natural death was imminent because of extreme pathology (causes revealed at necropsy include renal failure, severely debilitating osteoarthritis, and severe diabetes).
Natural populations of East Africa.
Reliable records of birth and death are only available for females in free-living populations: whereas females remain in their natal social groups, males disperse from their natal groups and are difficult to follow. We present age-specific mortality data for all females, and fit models to mortality data for adult females from two free-living populations, one in and around Amboseli National Park, Kenya (P. hamadryas cynocephalus), the other in Gombe National Park, Tanzania (P. hamadryas anubis) (18–21). Menarche in baboons occurs between 4 and 5 years of age in wild populations and at about 3.5 years at SFBR; here, we treat as adults all females 5 years of age and older. In both populations, study groups are monitored every 2–3 days on average, and all demographic events are recorded. In Amboseli, 274 females contributed to the vital rate data, 120 of whom were adults. As of 31 December 1999, 184 had died and 90 were alive (right censored). In Gombe, 399 females contributed to the data, 198 of whom were adults. As of 31 January 2000, 225 had died, and 174 were alive. The two populations are similar in social structure and behavior, but differ in ecological parameters. Baboons in Gombe experience no predation and a rich habitat that, for some social groups in some years was augmented by raiding human harvests of fish. Amboseli baboons experience substantial predation and the groups included in this analysis subsist entirely on wild foods in a semiarid habitat. In both populations, the original study groups (two in Amboseli and three in Gombe) fissioned during the study period to produce new social groups (four in Amboseli and eight in Gombe). Within each population, data from all study groups and their fission products were pooled for demographic analysis, to produce one set of vital rates for each population.
Demographic Analysis.
Life tables for females of all ages of SFBR, Gombe, and Amboseli were constructed with the actuarial life table method executed in the sas lifetest procedure (SAS Institute, Cary, NC). Age-specific mortality was estimated at the midpoint of 1-year intervals by counting the number of deaths and censored observations in each interval (i.e., median death rate in x − 1 to x) as mx = 2qx/(2 − qx), where qx is the conditional probability of death from age x − 1 to x.
Parametric models of adult mortality were fit with maximum likelihood methods executed in winmodest (23) in a two-step process. Only adult mortality was modeled in this analysis because intrinsic patterns of senescence will not be expressed in pre-adult life stages. First, for each population, goodness of fit was determined for the Logistic (μx = λeγx[1 + (λs/γ )(eγx − 1)]−1) and for the nested case when s = 0, the Gompertz (μx = λeγx); λ is the baseline mortality at age 5 years, γ is the rate of exponential increase in adult mortality with age, and s is the deceleration rate of mortality increase. Based on likelihood tests, the Gompertz model (s = 0) was never rejected in favor of the Logistic model (s ≠ 0); thus, we do not consider the Logistic model further. Second, a likelihood ratio test was used to compare the parameters γ and λ among populations. For each pair of populations the full model (each population with independent λ and γ) was compared by likelihood ratio tests to models that assume common γ, common λ, or common λ and γ (24). The MRDT was computed as (ln2)/γ, as in ref. 12. To compare rates of adult aging in female baboons and humans we analyzed vital rates from the U.S. female National Center for Health Statistics model life table based on 1997 death rates (10). Age classes 15–80 were used, as above, to test mortality model goodness-of-fit and to estimate parameters.
Results
Conditional on survival to adulthood (i.e., age 5), females of SFBR have a residual life expectancy of 16 years. An average adult female at SFBR lives to 21 years of age and record longevity is 33 years of age. In Amboseli, the residual life expectancy of 5-year-old females is 7.1 years, an average adult female lives to 12.1 years, and record longevity is 27 years of age. Gombe 5-year-old females have a residual life expectancy of 14.7 years, average adult survival to about 19.7 years, and record longevity is 27 years. For baboon life tables, see Table 2, which is published as supporting information on the PNAS web site, www.pnas.org. U.S. females, conditional on survival to age 15 years, live on average to 80 years of age (races combined; ref. 10).
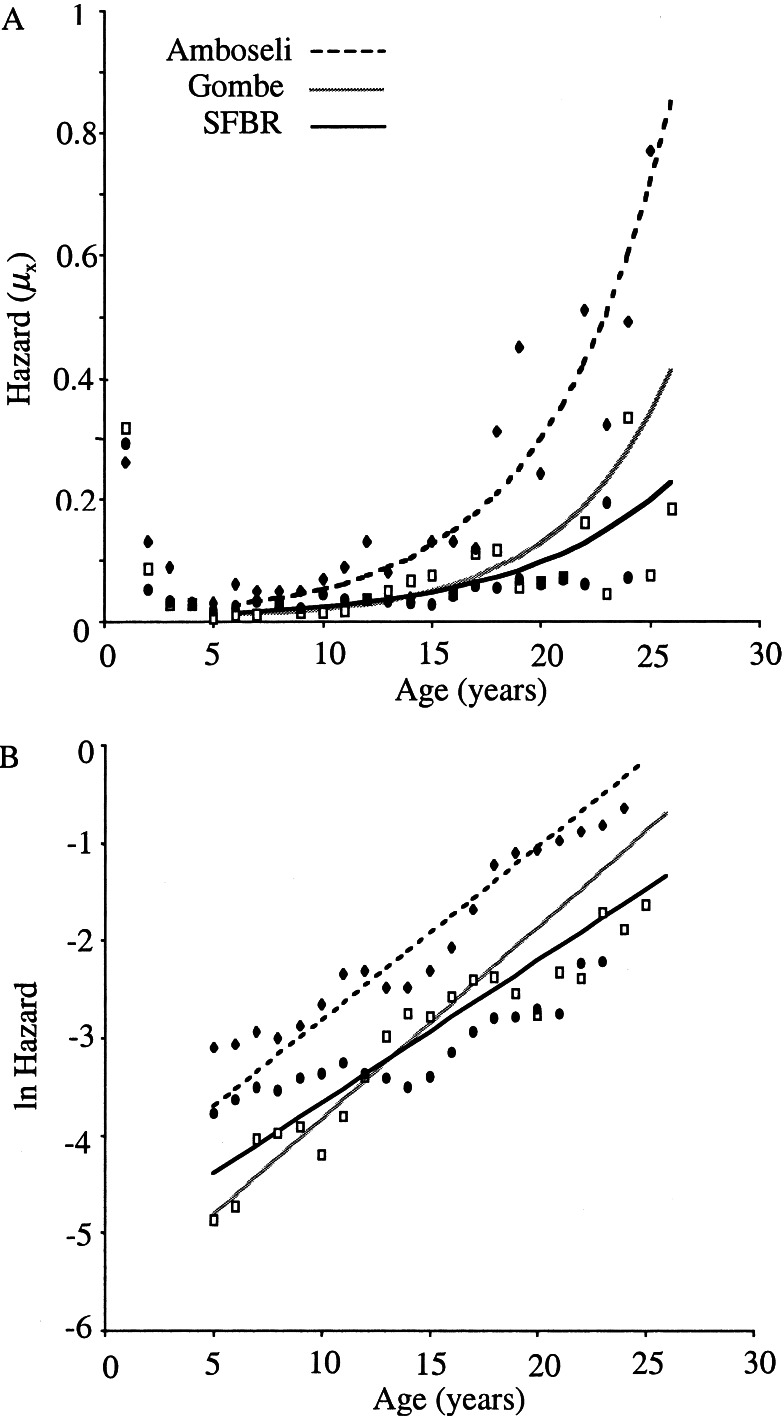
For female baboons in each population, the adult mortality was best
modeled by the Gompertz function (starting at age 5; Fig.
1A, Table
1). The Gompertz rate of adult mortality
change (γ) was relatively homogenous among the three baboon
populations (Table 1). A somewhat slower rate occurred in SFBR relative
to Gombe (χ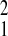 = 7.60, P = 0.006); no
other comparisons of this parameter supported significantly different
values (SFBR relative to Amboseli, χ
= 7.60, P = 0.006); no
other comparisons of this parameter supported significantly different
values (SFBR relative to Amboseli, χ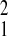 = 2.46,
P = 0.12; Gombe relative to Amboseli,
χ
= 2.46,
P = 0.12; Gombe relative to Amboseli,
χ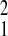 = 0.58, P = 0.45). In contrast,
the magnitude of the Gompertz baseline mortality parameter, λ, varied
markedly among populations. Amboseli had the highest level of initial
adult mortality (Amboseli relative to SFBR, χ
= 0.58, P = 0.45). In contrast,
the magnitude of the Gompertz baseline mortality parameter, λ, varied
markedly among populations. Amboseli had the highest level of initial
adult mortality (Amboseli relative to SFBR, χ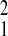 =
3.72; P = 0.05; Amboseli relative to Gombe,
χ
=
3.72; P = 0.05; Amboseli relative to Gombe,
χ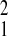 = 9.06, P = 0.003). SFBR had an
intermediate level of initial adult mortality; the lowest value was for
Gombe (SFBR relative to Gombe, χ
= 9.06, P = 0.003). SFBR had an
intermediate level of initial adult mortality; the lowest value was for
Gombe (SFBR relative to Gombe, χ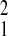 = 5.38,
P = 0.02). The relative magnitude of variation among
these parameters is evident on the log-mortality plot (Fig.
1B), where the fit models are largely parallel. The apparent
lack of fit between point estimates and parametric model in the case of
SFBR is misleading because trends on point estimates are inherently
biased (25). Thus, the three populations have similar rates of
mortality acceleration, with SFBR being slightly slower (MRDT are 3.9
and 3.5 years, respectively, for Amboseli and Gombe, and 4.8 years for
SFBR), but markedly different levels of baseline mortality that varied
3-fold from the lowest to the highest value.
= 5.38,
P = 0.02). The relative magnitude of variation among
these parameters is evident on the log-mortality plot (Fig.
1B), where the fit models are largely parallel. The apparent
lack of fit between point estimates and parametric model in the case of
SFBR is misleading because trends on point estimates are inherently
biased (25). Thus, the three populations have similar rates of
mortality acceleration, with SFBR being slightly slower (MRDT are 3.9
and 3.5 years, respectively, for Amboseli and Gombe, and 4.8 years for
SFBR), but markedly different levels of baseline mortality that varied
3-fold from the lowest to the highest value.
Figure 1.
(A) Mortality rates (hazards) for wild populations of Amboseli (⧫) and Gombe (□), and for the captive population of SFBR (●). (B) Natural logarithm of mortality rates. Points are smoothed log mortality based on the running average of mortality across a 3-year window. In both figures, adult mortality trajectories (lines) based on Gompertz parameters estimated from age 5 years.
Table 1.
Maximum likelihood estimates of Gompertz parameters, 95% confidence intervals, and mortality rate doubling estimates
| Population | Baseline mortality, λ | [λ: 95% CI] | Demographic rate of aging, γ | [γ: 95% CI] | MRDT, years |
|---|---|---|---|---|---|
| Baboon | |||||
| Gombe females | 0.00247 | [0.00133, 0.00456] | 0.1964 | [0.1658, 0.2327] | 3.5 |
| Amboseli females | 0.00843 | [0.00490, 0.01449] | 0.1774 | [0.1440, 0.2185] | 3.9 |
| SFBR females | 0.00510 | [0.00412, 0.00631] | 0.1457 | [0.1304, 0.1628] | 4.8 |
| SFBR males | 0.00651 | [0.00490, 0.00864] | 0.1691 | [0.1487, 0.1923] | 4.1 |
| Human | |||||
| U.S. female | 0.00004 | [0.00004, 0.00004] | 0.08691 | [0.08606, 0.08776] | 8.0 |
Baseline mortality (λ) is calculated from the interval at age 5 through the end of life in baboons, and from ages 15–80 in humans. The demographic rate of aging, γ, is the exponential parameter for mortality acceleration. MRDT is given by (ln2)/γ.
As with the baboons, the Logistic function did not provide a significantly improved fit relative to the Gompertz for U.S. females (based on the 1997 model life table, age 15–80 years, χ2 = 0.00086, P = 0.98). From the model table, baseline mortality (λ) at 15 years was 0.00004 and adult mortality rate increase (γ) through 80 years was 0.08691, which yields MRDT = 8.0. Finch et al. (12) reported human baseline mortality (U.S. females, 1980) as 0.0002 (at age 18 years) and γ = 0.0866 (MRDT = 8).
Discussion
Captive baboons at SFBR and wild baboons of East Africa demonstrate demographic senescence characterized by exponentially increasing mortality rate with age in adulthood. Mortality rate deceleration at advanced ages was not detected in these populations and the Gompertz mortality model provided the best way to parametrically characterize the trajectory of adult demographic aging. These data lack sufficient power to robustly test for deceleration among the oldest old, but it may be the case that deceleration is only a property of primate mortality trajectories when adults survive long into postreproductive ages, as occurs in humans but not in baboon.
Based on the estimates of Gompertz γ, mortality rate of SFBR adults doubles each 4.8 years, whereas mortality rate doubling time is 3.5 years for free-living adult female baboons in Gombe and 3.9 years for adult females in Amboseli. The variability among populations in their rate of demographic aging can be described by the coefficient of variance among estimates of γ, which is 16.0% (with sample size correction). Finch (12, 13), in reference to humans, noted this tendency for relative homogeneity among demographic rates of human aging over the 19th and 20th centuries, despite markedly disparate environmental experiences; the MRDT of humans ranged between 7 and 8.5 years. The extension of this pattern to a second primate species may suggest that the rate of demographic aging (γ) is a species-specific trait; extensive life tables from multiple wild populations of other taxa are required to evaluate this hypothesis. Relatively small differences among baboon populations in MRDT versus the larger difference documented here for baboon relative to human, indicate that genetic factors affecting the demographic rate of senescence exist, but may be fixed among taxonomic groups. Life expectancy, in contrast, is not a species characteristic because λ can vary markedly, as we observe here for three baboon populations, and as Finch has noted for humans. The coefficient of variance for λ among our sample of baboon populations is 60.7%, nearly four times larger than the variation observed for γ. The observed relative homogeneity of γ among baboons also implies that aging is an intrinsic feature of wild populations, consistent with a recent shift in conventional wisdom (26). In free-living populations, the high risks of disease, predation, and deprivation are expected to produce high levels of age-independent adult mortality such that senescence contributes little to realized adult life expectancy. However, among baboons, not only does aging occur in nature, the demographic rate of senescence observed in a captive population is recapitulated in two independent wild populations.
Why does baseline mortality λ vary among baboon populations? Diet, disease exposure, thermal and hydration stress, trauma, and deprivation during development are likely to impact individual vulnerability and to shape the distribution of frailty within populations. Gombe is located along the shore of Lake Tanganyika and contains rich areas of evergreen forest; food resources are relatively abundant, and the park is unusual in that natural vertebrate predators of baboon are largely absent (21). Gombe's salubrious environment may reduce the population's frailty, thus enhancing the similarity with the SFBR baboons. Gombe contrasts sharply with Amboseli, where predation is common and groups may spend up to 76% of the active day foraging and moving between widely spaced food patches, particularly in dry years (27). At SFBR, medical intervention and herd-management practices foster high infant survival rates, which may somewhat increase the prevalence of frail individuals in adult populations. Especially at the time when individuals graduate to adult social cages, frail individuals could elevate mean mortality rates among young adults. Thus, environmental factors are likely to generate heterogeneity in frailty among baboon populations.
Genetic factors may also contribute to population differences in frailty. Among chromosome extraction strains of Drosophila melanogaster, genetic variance for overall levels of mortality is produced primarily by differences in the Gompertz parameter λ (28). Although significant additive genetic variance occurs for both γ and λ, the additive genetic coefficient of variation (29) of λ (64%) is 3.5 fold larger than that of γ (18%). Thus, differences in mortality produced by the Gompertz parameter (γ) or by the mortality deceleration parameter (s of the Logistic mortality model) make minor contributions to differences in cohort life expectancy (30). How variance in mortality parameters is partitioned among populations suggests an important hypothesis concerning differences among individuals with respect to senescence. If γ is largely invariant among populations of a species, individuals within a population may also be relatively homogeneous in their personal rates of aging. Consequently, individual differences in mortality risk and lifespan may derive primarily from differences in frailty, which can be generated both by genetic and environmental sources. In this case, even if the correlation of parent and offspring frailty is high, heritability of lifespan will be modest (31).
The predominance in both Homo and Papio for heterogeneity of baseline mortality (λ) among populations or cohorts suggests that differences in frailty are the primary cause of variance in individual mortality risk within species. This conclusion is consistent with the observation that heritability of lifespan is modest for both genera [0.23–0.26 for Homo (3); 0.23 for Papio¶¶] and with the observation that frailty itself shows substantial heritability (h2 = 0.50) when estimated from Danish twins (32). Although longevity may be determined by direct genetic transmission within populations, it seems likely that factors for lifespan revealed through genomic mapping will reflect genes that alter underlying vulnerability at different ages rather than genes that directly determine the age of death. The identification of frailty-associated genes and their function could lead to both improved lifespan and health span of aging individuals. With the baboon of SFBR, genetic mapping of frailty and senescence determinants is feasible because human syntenic microsatellite markers have been characterized for the pedigreed colony and are used to conduct multipoint linkage analysis of complex traits (22, 33).
The replicate life tables of baboon provide a robust basis for comparative analysis of primate aging. In contrast to the conclusion of Finch et al. (12), where MRDT for Rhesus monkey was reported to be 15 years, we find that humans age at a slow demographic rate relative to at least one non-human primate, Papio. The intrinsic or typical life expectancy of any primate, however, remains an elusive characteristic because baseline mortality varies widely among populations within genera. To fully address questions on the nature of past human life expectancy, current postreproductive lifespan, and the prospects for future human longevity, we must discover why cohorts differ primarily in frailty and how the environment and genes shape this plastic demographic parameter of aging.
Supplementary Material
Acknowledgments
For permission to monitor the Amboseli baboon population, we are grateful to the Office of the President of Kenya, the Kenya Wildlife Service, and the members of the pastoralist communities of Amboseli and Longido. We thank D. A. Collins and the support of the Jane Goodall Institute U.S. for collection of the long-term data at Gombe Stream Research Center. Data collection on the Amboseli baboon population was made possible by the Chicago Zoological Society, National Science Foundation Grant IBN-9985910, and predecessors to J. Altmann, and depended on the help of numerous contributors to the Amboseli long-term database. A.M.B. was supported by National Institutes of Health Grant AG-05784. Work at Southwest Foundation for Biomedical Research and the Southwest Regional Primate Research Center, and the work of K.D.C., was supported by National Institutes of Health Grants HL-28972, U10-52636, and P51-RR1-13986. M.T. was supported by the Ellison Medical Foundation and National Institutes of Health Grant AG-16632.
Abbreviations
- MRDT
mortality rate doubling time
- SFBR
Southwest Foundation for Biomedical Research
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Bronikowski, A. M., Martin, L. J., Comuzzie, A. G., Mahaney, M. C., Packer, C., Carey, K. D. & Tatar, M. (2001) J. Am. Aging Assoc. 24, 115 (abstr.).
References
- 1.Vaupel J W. Philos Trans Biol Sci. 1997;352:1799–1804. doi: 10.1098/rstb.1997.0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cournil A, Kirkwood T B L. Trends Genet. 2001;17:233–235. doi: 10.1016/s0168-9525(01)02306-x. [DOI] [PubMed] [Google Scholar]
- 3.Herskind A M, McGue M, Hom N V, Sorensen T I A, Harvald B, Vaupel J W. Hum Genet. 1996;97:319–323. doi: 10.1007/BF02185763. [DOI] [PubMed] [Google Scholar]
- 4.Puca A A, Daly M J, Brewster S J, Matise T C, Barrett J, Shea-Drinkwater M, Kang S, Joyce E, Nicoli J, Benson E, et al. Proc Natl Acad Sci USA. 2001;98:505–508. doi: 10.1073/pnas.181337598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paine R R. Am J Phys Anthropol. 1989;79:51–61. doi: 10.1002/ajpa.1330790106. [DOI] [PubMed] [Google Scholar]
- 6.Acsadi G, Nemeskeri J. History of Human Life Span and Mortality. Budapest: Akademiai Kiado; 1970. [Google Scholar]
- 7.Wilmoth J R. Exp Gerontol. 2000;35:1111–1129. doi: 10.1016/s0531-5565(00)00194-7. [DOI] [PubMed] [Google Scholar]
- 8.Howell N. Demography of the Dobe !Kung. New York: de Gruyter; 2000. [Google Scholar]
- 9.Hill K, Hurtado A M. Ache Life History: The Ecology and Demography of a Foraging People. New York: de Gruyter; 1996. [Google Scholar]
- 10.Anderson R N. National Vital Statistics Reports 47. Hyattsville, MD: National Center for Health Statistics; 1999. [PubMed] [Google Scholar]
- 11.Judge D S, Carey J R. J Gerontol. 2000;55A:B201–B209. doi: 10.1093/gerona/55.4.b201. [DOI] [PubMed] [Google Scholar]
- 12.Finch C E, Pike M C, Witten M. Science. 1990;249:902–905. doi: 10.1126/science.2392680. [DOI] [PubMed] [Google Scholar]
- 13.Finch C E. Longevity, Senescence and the Genome. Chicago: Univ. of Chicago Press; 1990. [Google Scholar]
- 14.Vaupel J W, Carey J R, Christensen K, Johnson T E, Yashin A I, Holm N V, Iachine I A, Kannisto V, Khazaeli A A, Liedo P, et al. Science. 1998;280:855–860. doi: 10.1126/science.280.5365.855. [DOI] [PubMed] [Google Scholar]
- 15.Vaupel J W, Manton K G, Stallard E. Demography. 1979;16:439–454. [PubMed] [Google Scholar]
- 16.Sacher G A. In: Handbook of the Biology of Aging. Finch C E, Hayflick L, editors. New York: Van Nostrand Reinhold; 1977. pp. 582–638. [Google Scholar]
- 17.Gage T B. Annu Rev Anthropol. 1998;27:107–221. doi: 10.1146/annurev.anthro.27.1.197. [DOI] [PubMed] [Google Scholar]
- 18.Packer C, Tatar M, Collins A. Nature (London) 1998;392:807–811. doi: 10.1038/33910. [DOI] [PubMed] [Google Scholar]
- 19.Altmann J, Altmann S, Hausfater G. In: Reproductive Success. Clutton-Brock T H, editor. Chicago: Univ. of Chicago Press; 1988. pp. 403–418. [Google Scholar]
- 20.Alberts S C, Altmann J. In: Primate Life Histories and Socioecology. Kappeler P, Pereira M, editors. Chicago: Univ. of Chicago Press; 2002. [Google Scholar]
- 21.Packer C, Collins D A, Eberly L E. Philos Trans R Soc London. 2000;355:1627–1635. doi: 10.1098/rstb.2000.0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin L J, Blangero J, Rogers J, Mahaney M C, Hixson J E, Carey K D, Comuzzie A G. Hum Biol. 2001;73:787–800. doi: 10.1353/hub.2001.0086. [DOI] [PubMed] [Google Scholar]
- 23.Pletcher S D. WINMODEST 1.0.2. London: University College; 1999. [Google Scholar]
- 24.Pletcher S D. J Evol Biol. 1999;12:430–439. [Google Scholar]
- 25.Promislow D E L, Tatar M, Pletcher S, Pletcher S, Carey J R. J Evol Biol. 1999;12:314–328. [Google Scholar]
- 26.Kirkwood T B, Austad S N. Nature (London) 2000;408:233–238. doi: 10.1038/35041682. [DOI] [PubMed] [Google Scholar]
- 27.Bronikowski A M, Altmann J. Behav Ecol Sociobiol. 1996;39:11–25. [Google Scholar]
- 28.Promislow D E L, Tatar M, Khazaeli A A, Curtsinger J W. Genetics. 1996;143:839–848. doi: 10.1093/genetics/143.2.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Houle D. Genetics. 1992;130:195–204. doi: 10.1093/genetics/130.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pletcher S D, Khazaeli A A, Cursinger J W. J Gerontol. 2000;55A:B381–B389. doi: 10.1093/gerona/55.8.b381. [DOI] [PubMed] [Google Scholar]
- 31.Vaupel J W. Demography. 1988;25:277–287. [PubMed] [Google Scholar]
- 32.Yashin A I, Iachine I A, Harris J R. Behav Genet. 1999;29:11–19. doi: 10.1023/a:1021481620934. [DOI] [PubMed] [Google Scholar]
- 33.Rogers J, Mahaney M C, Witte S M, Nair S, Newman D, Wedel S, Rodriques L A, Rice K S, Slifer S H, Perelygin A, et al. Genomics. 2000;67:237–247. doi: 10.1006/geno.2000.6245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.