Polymers of actin (F-actin) form an integral part of the structural framework that supports the plasma membrane of our cells while providing a platform for signaling and metabolic proteins. Most subunits in an actin filament hydrolyze a single molecule of ATP to ADP over the F-actin's lifetime. This hydrolysis is the critical timekeeper of F-actin longevity that informs a host of accessory proteins about the state of the filament (1). Here, we discuss the structural changes within each subunit of F-actin that are induced by the nucleotide hydrolysis.
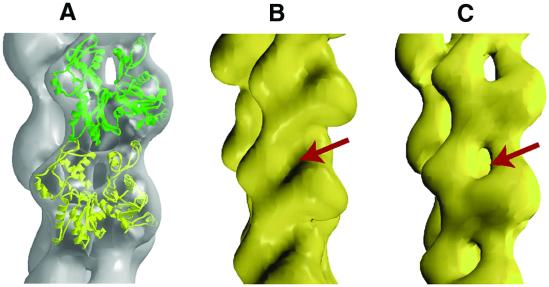
The tendency of monomeric actin to form polymers of varying lengths has so far prevented crystallization and atomic resolution structural analysis of F-actin. Previously, crystal structures of monomeric ATP-bound actin were used to construct a helical model of F-actin (2). Recently, the structure of tetramethylrhodamine-5-maleimide (TMR)-labeled ADP-bound actin monomer was determined (3) and proposed (4) to represent the ADP conformation of protomers in actin filaments. A structural change in the DNaseI-binding loop from a random coil to a helix in the ADP-bound actin (3) was suggested (4) to explain how some regulatory proteins discriminate between ADP- and ATP-actin filaments. However, fitting this structure of the ADP-bound actin monomer into electron microscopic reconstructions of ADP-bound actin filaments shows discrepancies. Differences are most prominent around the nucleotide-binding cleft (Fig. 1, red arrow). It is more open in the protomers of ADP-bound F-actin filaments (Fig. 1C) compared with ADP-bound TMR-labeled actin (Fig. 1B).
Fig 1.
(A) The ADP-bound TMR-labeled actin structure (yellow and green) docked into the electron microscopic reconstruction (gray in A and yellow in C) of the ADP-bound F-actin. (B) Rendered surface of the docked ADP-bound TMR-labeled structure. The surface has been rendered at ≈22 Å to match the resolution of the electron microscopic data. (C) Electron microscopic reconstruction of F-actin in ADP state. Arrows point to the closed (B) and open (C) nucleotide-binding cleft that is apparent in these low-resolution surfaces.
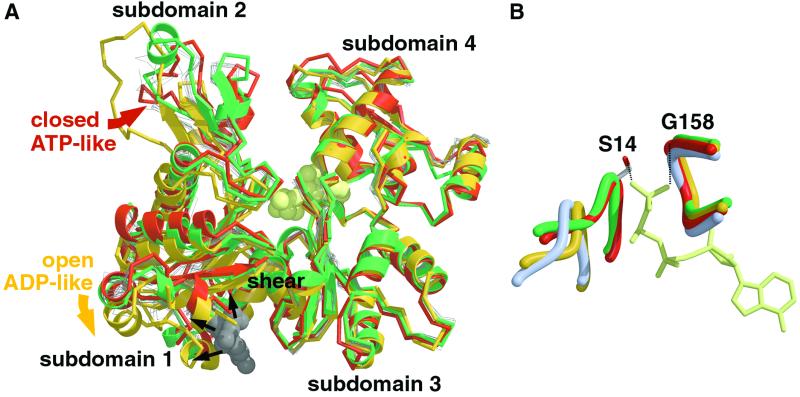
What then is the structure of the actin monomer in the ADP state? A different model for nucleotide-induced conformational changes in F-actin arises from electron microscopic data and comparison of atomic-resolution structures of actin. All available crystal structures of actin fall into two distinct categories (Fig. 2A): the “closed” group, which includes the closed structure of actin found in one actin-profilin complex (ref. 5; Fig. 2A, red), and the “open” group, including actin in an open conformation (Fig. 2A, yellow), found in a different actin-profilin complex (6). Previous analysis of these profilin-bound actin structures suggested that closing or opening of the nucleotide-binding cleft occurred through rotation of the two actin domains (subdomains 1 plus 2 and 3 plus 4, respectively) around the extended hinge (or shear, Fig. 2A, marked) area of actin (7). Our comparison of 12 actin structures supports this proposal and suggests a general mechanism for controlling movements of actin subdomains between the open and closed conformations of actin. This mechanism is similar to that used by other families of nucleotide-hydrolyzing proteins (NTPases).
Fig 2.
Comparative analysis of actin structures. (A) Superposition of available crystal structures of actin. The superposition was done by using coordinates of Cα atoms of residues from subdomains 3 (amino acids 150–180 and 274–337) and 4 (amino acids 181–273) in each case. The closed and open actin structures (5, 6) from the actin-profilin complexes are shown in red and yellow, respectively. The ADP-bound TMR-labeled structure of the actin (3) is shown in green, with TMR drawn as a gray space-filling model. The polypeptide chains of the rest of available actin structures are drawn as thin gray lines. The position of bound nucleotide (ATP) is indicated by a space-filling model in cream. Subdomains 1–4 of actin are indicated, and their movements between the open and closed conformations are shown by red and yellow arrows, respectively. The black arrows indicate steric clashes, which would occur between the open actin structure and the intercalated TMR. (B) Conformations of the γ-phosphate-sensing loops in closed (red) and open (yellow) structures of actin. The closed conformations of the nucleotide-sensing loops are defined by the γ-phosphate of the bound nucleotide (in cream), which coordinates two conserved residues, Ser-14 and Gly-158. Hydrogen bonds are indicated by dashed lines. The closed conformation of the nucleotide cleft is stabilized further by a network of interactions that involve residues from all four subdomains of actin. The closed and open conformations of the nucleotide-binding loops of the TMR-labeled actin (3) and Arp3 (11) are shown in green and blue, respectively.
Various families of NTPases employ a so-called “γ-phosphate sensing” mechanism to change their molecular conformations between different nucleotide states (8, 9). In the triphosphate state (ATP or GTP) of these proteins, conserved Gly and Ser or Thr residues in loops surrounding the nucleotide form hydrogen bonds with the γ-phosphate, a bridge that keeps the nucleotide pocket closed. In the diphosphate state (ADP or GDP), this bridge disengages and the nucleotide-binding cleft opens.
Comparing the nucleotide-binding loops of the open and closed actin structures reveals that the general features of the γ-phosphate-sensing mechanism are also present in actin. A conserved glycine (Gly-158) and serine (Ser-14) from two invariant nucleotide-binding loops (Fig. 2B, red) interact with the γ-phosphate of ATP (Fig. 2B, cream) in the closed state of actin. A network of side chain and main chain interactions transmits the positions of these loops to the rest of the actin structure, bringing the two domains of actin together in its closed conformation (Fig. 2A, B red). In the open conformation of actin, the γ-phosphate-binding loops are disengaged (Fig. 2B, yellow), causing the two domains to move apart and open the nucleotide-binding cleft (Fig. 2A, yellow).
The apparent similarities between NTPases and actin suggest that these closed and open conformations may represent the ATP/ADP.Pi and the ADP/no nucleotide states of actin, respectively. This proposal is consistent with the results of docking the closed and open structures of actin into electron microscopic reconstructions of the ADP.Pi-like and ADP-bound forms of F-actin (10). Remarkably, the recently solved crystal structures of nucleotide-free actin-like proteins Arp2 and Arp3 (11) revealed an open conformation similar to that observed in the open structure of actin. Furthermore, the conformations of the phosphate-binding loops of the nucleotide-free Arp3 (Fig. 2B, blue; part of the Arp2 structure is not resolved and, therefore, not used for the illustration) are identical to those in the open actin structure. It was hypothesized that the binding of ATP could close the nucleotide-binding cleft of these proteins (11).
Interestingly, conformations of the nucleotide-binding loops and the positions of the actin subdomains in the ADP-bound TMR-labeled actin (3) place this structure in the closed ATP-like category (Fig. 2 A and B, green). This observation is explained by the attached TMR (Fig. 2A, gray), which intercalates between subdomains 1 and 3 in the shear region and stabilizes the closed conformation by encouraging the actin's domains to move and the nucleotide cleft to close, as TMR cannot fit into the open structure. We propose that this intercalating TMR uncouples the γ-phosphate sensing mechanism from the structure of the actin cleft so that ADP is found in a closed structure. Similar uncoupling has been observed (12) in the ADP- and ATP-bound structures of actin complexed with DNase I, as well as in the structures of other NTPases (reviewed in refs. 9 and 13). In all cases, small free-energy differences relate the closed ATP and open ADP conformations. Because of the small energy barrier, external factors (crystallization conditions, crystal lattice interactions, the presence of bound ligands, etc.) affected the equilibrium between these conformations.
It is important to note that within both the closed and open categories of actin the structural conformations of subdomain 2 vary (Fig. 2A). The poorly structured features of subdomain 2 seem to be designed to accommodate varied binding interactions with other proteins. In the absence of specific binding partners, loops of subdomain 2 could adopt stochastic conformations so that even crystal contacts could alter them. The DNase I-binding loop of subdomain 2 in TMR-labeled actin crystals is packed against three helices (amino acid residues 80–93, 223–230, and 252–262, respectively; see Fig. 3, which is published as supporting information on the PNAS web site, www.pnas.org) from two neighboring actin symmetry molecules (3) that probably trigger helix formation in this loop. However, none of these helices is located near the DNase I-binding loop in the actin filament (2), suggesting that the formation of a helix by this loop does not represent a structural determinant for discrimination between the ATP and ADP states of actin filament.
What then is the structural basis for discrimination between different nucleotide states of actin by various actin-binding proteins that regulate the dynamic process of actin assembly and disassembly in vivo? Coupling the γ-phosphate sensing to the opening and closing of the nucleotide-binding cleft offers an elegant solution to this long-standing problem. We suggest that the γ-phosphate-controlled movements of actin domains that accompany closing and opening of the nucleotide-binding cleft provide a structural basis for this discrimination. After phosphate release, the rotational movements of actin domains would change longitudinal inter-subunit contacts in F-actin (10, 14). The resulting changes in the accessible surface would open specific docking sites (15) for the ADF/cofilin/destrin superfamily of depolymerizing proteins. In the closed ATP/ADP.Pi state of F-actin, part of this binding interface is not accessible.
Our proposed mechanism for discriminating between the ADP and ATP-like states of actin predicts that the binding of DNase I (12), which clamps subdomains 2 and 4 at the top of the nucleotide pocket, stabilizes the closed ATP-like conformation of actin. Segment-1 of gelsolin (GS-1) would stabilize the closed conformation of actin by intercalating between subdomains 1 and 3 in the shear area of actin, requiring actin's domains to rotate and to close the nucleotide cleft (J.F.D., E.P.S., J.A.S., and R.J.F., data not shown). In contrast, the binding of profilin seems to be compatible with both the closed (5) and open (6) conformations of actin. This observation is most likely related to the cellular function of profilin, which accelerates the exchange of ADP for ATP while remaining bound to actin in both states.
In summary, based on the combined electron microscopic and crystallographic data, we propose a model for the dynamics of F-actin assembly and its regulation (Fig. 4, which is published as supporting information on the PNAS web site). This model combines the helical structure of F-actin (2) with conformational transitions between the closed and open states of actin (7). By analogy with other NTPases, we assign the closed and open conformations of actin to the functionally critical ATP/ADP.Pi and ADP/no nucleotide states of actin, respectively. We posit that the conformational transitions between the closed and open states of actin are nucleotide-dependent and controlled by a γ-phosphate-sensing mechanism that seems to be similar in various NTPases. By employing this mechanism, nature has designed a “cytoskeletal timekeeper” that controls dynamics of actin assembly by keeping the newly formed (ATP/ADP.Pi-bound) actin filaments stable while marking the older ones (ADP-bound) for depolymerization by regulatory actin-binding proteins (1).
Supplementary Material
Acknowledgments
This work was supported by funding from a National Institutes of Health program project grant. J.F.D. was supported by a postdoctoral fellowship from the Natural Sciences and Engineering Research Council of Canada.
References
- 1.Pollard T. D., Blanchoin, L. & Mullins, R. D. (2000) Annu. Rev. Biophys. Biomol. Struct. 29, 545-576. [DOI] [PubMed] [Google Scholar]
- 2.Holmes K. C., Popp, D., Gebhard, W. & Kabsch, W. (1990) Nature (London) 347, 44-49. [DOI] [PubMed] [Google Scholar]
- 3.Otterbein L. R., Graceffa, P. & Dominguez, R. (2001) Science 293, 708-711. [DOI] [PubMed] [Google Scholar]
- 4.De La Cruz E. M. & Pollard, T. (2001) Science 293, 616-618. [DOI] [PubMed] [Google Scholar]
- 5.Schutt C. E., Myslik, J. C., Rozycki, M. D., Goonesekere, N. C. W. & Lindberg, U. (1993) Nature (London) 365, 810-816. [DOI] [PubMed] [Google Scholar]
- 6.Chik J. K., Lindberg, U. & Schutt, C. E. (1996) J. Mol. Biol. 263, 607-623. [DOI] [PubMed] [Google Scholar]
- 7.Page R., Lindberg, U. & Schutt, C. E. (1998) J. Mol. Biol. 280, 463-474. [DOI] [PubMed] [Google Scholar]
- 8.Vetter I. R. & Wittinghofer, A. (2001) Science 294, 1299-1304. [DOI] [PubMed] [Google Scholar]
- 9.Sablin E. P. & Fletterick, R. J. (2001) Curr. Opin. Struct. Biol. 11, 716-724. [DOI] [PubMed] [Google Scholar]
- 10.Belmont L. D., Orlova, A., Drubin, D. G. & Egelman, E. H. (1999) Proc. Natl. Acad. Sci. USA 96, 29-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson R. C., Turbedsky, K., Kaiser, D. A., Marchand, J.-B., Higgs, H. N., Choe, S. & Pollard, T. D. (2001) Science 294, 1679-1684. [DOI] [PubMed] [Google Scholar]
- 12.Kabsch W., Mannherz, H. G., Suck, D., Pai, E. F. & Holmes, K. C. (1990) Nature (London) 347, 37-44. [DOI] [PubMed] [Google Scholar]
- 13.Geeves M. A. & Holmes, K. C. (1999) Annu. Rev. Biochem. 68, 687-728. [DOI] [PubMed] [Google Scholar]
- 14.Galkin V. E., VanLoock, M. S., Orlova, A. & Egelman, E. H. (2002) Curr. Biol. 12, 570-575. [DOI] [PubMed] [Google Scholar]
- 15.Galkin V. E., Orlova, A., Lukoyanova, N., Wriggers, W. & Egelman, E. H. (2001) J. Cell Biol. 153, 75-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.