Abstract
The light-harvesting properties of cationic conjugated polymers are used to sensitize the emission of a dye on a specific peptide nucleic acid (PNA) sequence for the purpose of homogeneous, “real-time” DNA detection. Signal transduction is controlled by hybridization of the neutral PNA probe and the negative DNA target. Electrostatic interactions bring the hybrid complex and cationic polymer within distances required for Förster energy transfer. Conjugated polymer excitation provides fluorescein emission >25 times higher than that obtained by exciting the dye, allowing detection of target DNA at concentrations of 10 pM with a standard fluorometer. A simple and highly sensitive assay with optical amplification that uses the improved hybridization behavior of PNA/DNA complexes is thus demonstrated.
Methods for DNA sequence identification in real time and with high sensitivity are of great scientific and economic interest (1–3). Their applications include medical diagnostics, identification of genetic mutations, gene delivery monitoring, and specific genomic techniques (4). Cationic organic dyes, such as ethidium bromide and thiazole orange, emit when intercalated into the grooves of double-stranded DNA and serve as direct DNA hybridization probes, but lack sequence specificity (5, 6). Energy/electron transfer chromophore pairs for strand-specific assays exist, but require chemical labeling of two nucleic acids or dual modification of the same altered strand (i.e., molecular beacons) (7, 8). Difficulties in labeling two DNA sites result in low yields, high costs, and singly labeled impurities, which lower detection sensitivity (9). Much of the motivation behind improving DNA sensing is to develop simple and economic methods for evaluating strand-specific hybridization that uses the ease of homogeneous fluorescence assays with minimal DNA modification and enhanced signal amplification.
Conjugated polymers (CPs) are characterized by a delocalized electronic structure and can be used as highly responsive optical reporters for chemical and biological targets (10, 11). Because the effective conjugation length is substantially shorter than the number of repeat units, the backbone serves to hold a series of conjugated segments in close proximity. Thus, CPs are efficient for light harvesting and enable optical amplification via Förster transfer (12). Water-soluble CPs show exceptional fluorescence quenching efficiencies in the presence of oppositely charged acceptors and are of particular interest for transduction of biological recognition events (13).
Spontaneous interpolymer complexation between cationic polyelectrolytes and DNA is known and is largely the result of cooperative electrostatic forces (14–16). Hydrophobic interactions between aromatic polymer units and DNA bases were also recently recognized (17). The free energy of polyelectrolyte/DNA interactions is controlled by the structure of the participating species, in conjunction with solution variables such as pH, ionic strength, and temperature (18). The strength and specificity of these interactions has recently been coordinated to recognize the tertiary structure of plasmid DNA (19).
The recent introduction of peptide nucleic acids (PNAs) opened the door for new research and diagnostic applications (20, 21). In PNAs, the negatively charged phosphate linkages in DNA are replaced with peptomimetic neutral amide linkages. PNA/DNA complexes form more quickly and are tighter and more specific than analogous DNA/DNA complexes (22). These properties are largely caused by the absence of the Coulombic repulsion found between negatively charged DNA strands. PNA complexes are thus more thermally stable and, by virtue of their backbone, less susceptible to biological degradation by nucleases, proteases, and peptidases (23, 24). Additionally, their general insensitivity to ionic strength and pH during hybridization provides a wider platform for DNA detection.
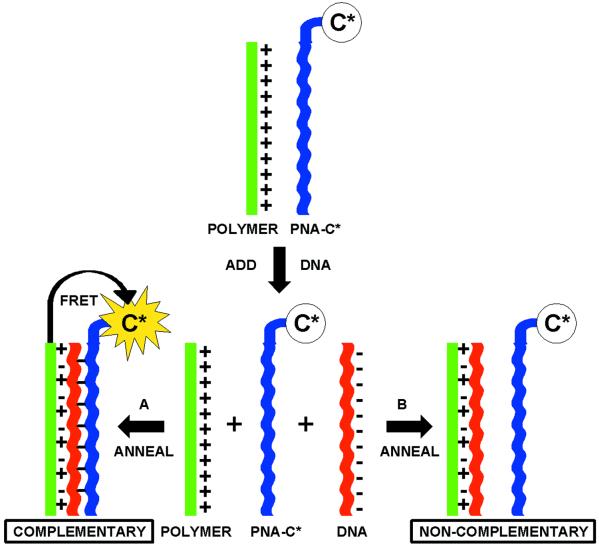
A scheme for detecting PNA/DNA interactions based on the considerations given above is shown in Scheme . Consider a solution that contains a cationic-conjugated polymer (CCP, shown in green) and a PNA strand (shown in blue) labeled with a chromophore dye (C*). The optical properties of the CCP and C* are chosen to favor Förster energy transfer (FRET) from CCP (donor) to C* (acceptor) (25). In the initial solution no electrostatic interactions are present, resulting in an average CCP–C* distance too large for effective FRET. Single-stranded DNA (ssDNA) is then added and an appropriate annealing protocol is followed. Situation A corresponds to addition of a complementary ssDNA (shown in red), which hybridizes with the target PNA. Hybridization endows the C*-bearing macromolecule with multiple negative charges. Electrostatic interactions should cause the formation of a complex and a decrease in the average CCP–C* distance, allowing for FRET. When a ssDNA that does not match the PNA sequence is added (shown in red), situation B, hybridization does not take place. Electrostatic complexation occurs only between the CCP and DNA while the CCP-PNA-C* distance remains too large for FRET. PNA/ssDNA hybridization is therefore measured by FRET efficiency, or the enhanced C* emission. The overall scheme serves as a probe for the presence of specific ssDNA sequences in solution.
Scheme 1.
Schematic representation for the use of a water-soluble CP with a specific PNA-C* optical reporter probe to detect a complementary ssDNA sequence.
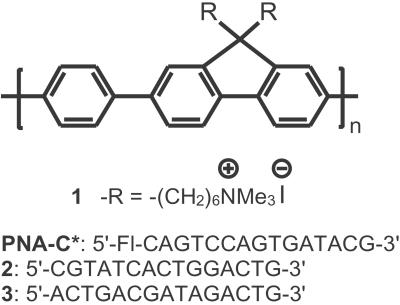
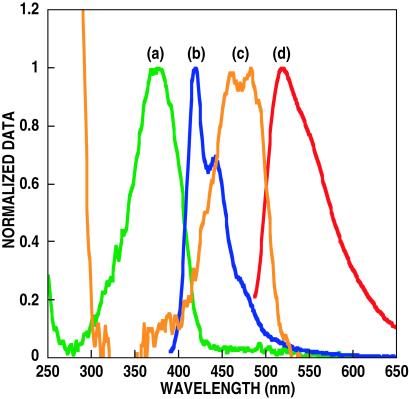
Scheme was tested by using the cationic water-soluble CP poly(9,9-bis(6′-N,N,N-trimethylammonium)-hexyl)-fluorene phenylene) containing iodide counteranions (1, in Scheme ) (26). A PNA probe corresponding to a CAGTCCAGTGATACG base sequence (in Scheme ) with fluorescein at the 5′ position of the strand was used as PNA-C* (blue fragment in Scheme ). The absorption and emission spectra of 1 and PNA-C* in Fig. 1 show an optical window for the excitation of 1, between the DNA, PNA, and C* absorptions. There is excellent overlap between the emission of 1 and the absorption of C* to ensure FRET.
Scheme 2.
Molecular structure of 1 and the PNA-C* and DNA sequences.
Fig 1.
Absorption [(a) green and (c) orange] and emission [(b) blue and (d) red] spectra of polymer 1 and single-stranded PNA probe 2, respectively. Fluorescence was measured by exciting at 380 and 480 nm, for 1 and 2, respectively.
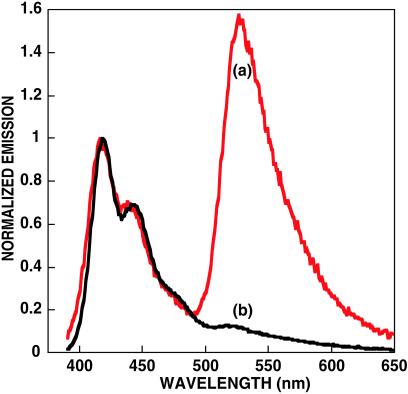
The PNA-C* probe ([PNA-C*] = 2.5 × 10−8 M) was annealed at 2°C below its Tm (72°C at 10−8 M, pH = 5.5) in the presence of an equimolar amount of its complementary 15-bp ssDNA, 2, and in an identical fashion with a noncomplementary 15-base ssDNA, 3 (27).‡ Annealing was accomplished in the absence of buffer, i.e., at low ionic strength, and the subsequent melting was monitored by UV/visible spectroscopy (28). Addition of 1 in water ([1] = 2.3 × 10−7 M) and comparison of the resulting fluorescence (Fig. 2) reveals a FRET ratio >11 times higher for the PNA/DNA hybrid, relative to the noncomplementary pair.§ These FRET differences demonstrate the validity of Scheme . Furthermore, the fluorescein emission is more than eight times more intense than that obtained from direct C* excitation in the absence of 1.¶ The increased C* emission in the energy transfer complex indicates the optical amplification provided by the CP. The sensitized acceptor emission is turned on by the addition of complementary DNA.
Fig 2.
Emission spectra of PNA-C* in the presence of complementary [(a) red] and noncomplementary [(b) black] DNA by excitation of polymer 1. Conditions are in water at pH = 5.5. The spectra are normalized with respect to the emission of polymer 1.
Energy transfer was optimized by varying the ratio of 1 to PNA-C*. At a concentration of [PNA-C*] = 2.5 × 10−8 M, initial additions of 1 cause an immediate rise in the FRET ratio. When [1] far exceeds [PNA-C*], a decrease is observed. The maximum in the FRET ratio corresponds to a near 1:1 ratio of polymer chains to PNA strands, according to previously published molecular weight information (Mn = 8,600 g/mol). Such a relationship should not be surprising, because when [1]/[PNA-C*] <1, not all ssDNA/PNA-C* hybrid strands can be complexed efficiently to independent polymer chains. Conversely, in the [1]/[PNA-C*] >1 regime, not all of the photons harnessed by 1 (the donor) can be transferred to the DNA/PNA-C* hybrid (the acceptor). Note that the C* emission at the saturation point is >25 times more intense than that obtained by direct C* excitation (480 nm), giving further evidence of signal amplification. This amplification allows detection of C* emission when [PNA-C*/DNA complex] = 10 pM with a standard fluorometer [PTI (South Brunswick, NJ) Quantum Master fluorometer equipped with a Xenon lamp excitation source and a Hamamatsu (Middlesex, NJ) photomultiplier tube].
Examination of Fig. 2 shows a small fluorescein signal from the nonhybridized PNA probe (situation B, Scheme ), which results from hydrophobic interactions between 1 and PNA-C* (29, 30). Solutions containing 10% ethanol, under the identical conditions as the experiments shown in Fig. 2, show a decrease in C* emission. The presence of the organic solvent decreases hydrophobic interactions and reduces C* emission by a factor of 3, at which point the signal is almost undetectable with a standard fluorometer.
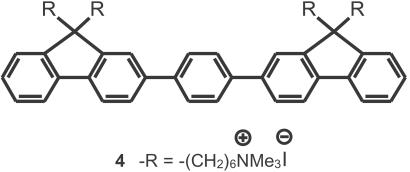
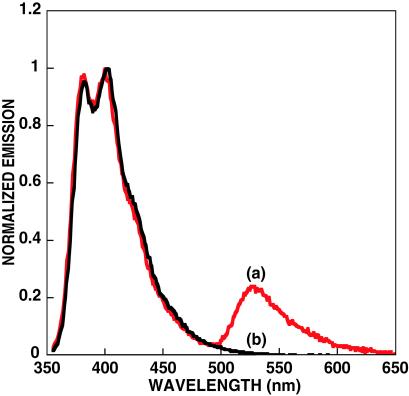
A water-soluble conjugated oligomer, (4, in Scheme ), of similar chemical structure to that of 1 was also examined within the context of Scheme . Although the smaller molecule will not display the same signal amplification, it is useful to deconvolute structure property relationships, which are difficult to determine with the inherent polydispersity and batch-to-batch variations found in polymers. Further, in aqueous media, 4 is considerably more soluble than 1, and hydrophobic interactions with neutral PNA should not be as severe. Fig. 3 ([4] = 6.7 × 10−8 M and [PNA-C*] = 2.5 × 10−8 M) shows C* emission only when the complementary ssDNA was present. Comparison of Figs. 2 and 3 implies that use of CPs with higher molecular weights will lead to higher FRET ratios. We thus anticipate that significantly higher sensitivity can be achieved.
Scheme 3.
Molecular structure of 4.
Fig 3.
Emission spectra of PNA-C* in the presence of complementary [(a) red] and noncomplementary [(b) black] DNA by excitation of 4. Conditions are in water and pH = 5.5. The spectra are normalized with respect to the emission of 4.
In summary, it is possible to take advantage of the optical amplification of CPs to detect DNA hybridization to a singly labeled PNA strand. This method provides a homogeneous assay that uses the ease of fluorescence detection methods and capitalizes on the enhanced hybridization behavior found in PNA–DNA interactions. As shown in Figs. 2 and 3, the reporter emission “turns on” only when the target ssDNA is present in solution. The overall strategy also eliminates the need for multiple probes and complex DNA structures. In a practical assay, donor emission could easily be filtered off and light intensity measured with a simple photodiode. The concept could also be used in PCR analysis or, because of the large signal amplification, as a stand-alone assay for the detection of a specific DNA sequence. Additionally, PNAs also have the ability to form triplex structures by binding to double-stranded DNA (dsDNA) and to invade a dsDNA sequence and displace the DNA strand of the same sequence (31–33). We expect that such interactions would allow the use of PNA-C*/CP sensor platforms to be used in direct dsDNA detection. Further optimization of CP structure/optical properties with a better understanding of the forces that control the association between conjugated polyelectrolytes, DNA, and PNA will yield superior detection platforms.
Acknowledgments
We thank the National Institutes of Health (GM62958-01), Office of Naval Research (N00014-98-1-0759), and National Science Foundation (DMR-0097611) for partial support of this work.
Abbreviations
PNA, peptide nucleic acid
ssDNA, single-stranded DNA
CP, conjugated polymer
CCP, cationic CP
C*, chromophore dye
FRET, fluorescence resonance energy transfer
An interactive version of ref. 27 can be found at Applied Biosystem's custom probe designer web site: www.appliedbiosystems.com/cgi-bin/calculator/ab_configured/oligodesigner/designer.cgi.
The FRET ratio is defined as the integrated acceptor emission over the integrated emission of the donor.
Fluorescein at pH = 5.5 is not in its high quantum yield dianionic form, thus we would expect higher C* emission at higher pH, but at the expense of charge neutrality on the PNA-C* complex.
References
- 1.Wang J. (2000) Nucleic Acids Res. 28, 3011-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Umek R. M., Lin, S. W., Vielmetter, J., Terbrueggen, R. H., Irvine, B., Yu, C. J., Kayyem, J. F., Yowanto, H., Blackburn, G. F., Farkas, D. H. & Chen, Y. P. (2001) J. Mol. Diag. 3, 74-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schork N. J., Fallin, D. & Lanchbury, J. S. (2000) Clin. Genet. 58, 250-264. [DOI] [PubMed] [Google Scholar]
- 4.Balakin K. V., Korshun, V. A., Mikhalev, I. I., Maleev, G. V., Malakhov, A. D., Prokhorenko, I. A. & Berlin, Y. A. (1998) Biosens. Bioelectron. 13, 771-778. [DOI] [PubMed] [Google Scholar]
- 5.LePecq J. B. & Paoletti, C. (1967) J. Mol. Biol. 27, 87-106. [DOI] [PubMed] [Google Scholar]
- 6.Petty J. T., Bordelon, J. A. & Robertson, M. E. (2000) J. Phys. Chem. B 104, 7221-7227. [Google Scholar]
- 7.Cardullo R. A., Agrawal, S., Flores, C., Zamechnik, P. C. & Wolf, D. E. (1988) Proc. Natl. Acad. Sci. USA 85, 8790-8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castro A. & Williams, J. G. K. (1997) Anal. Chem. 69, 3915-3920. [DOI] [PubMed] [Google Scholar]
- 9.Knemeyer J., Marméloch, N. & Sauer, M. (2000) Anal. Chem. 7, 3717-3724. [DOI] [PubMed] [Google Scholar]
- 10.McQuade D. T., Pullen, A. E. & Swager, T. M. (2000) Chem. Rev. 100, 2537-2574. [DOI] [PubMed] [Google Scholar]
- 11.Chen L., McBranch, D. W., Wang, H.-L., Helgeson, R., Wudl, F. & Whitten, D. G. (1999) Proc. Natl. Acad. Sci. USA 96, 12287-12292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dogariu A., Gupta, R., Heeger, A. J. & Wang, H. (1999) Synth. Metals 100, 95-100. [Google Scholar]
- 13.Wang J., Wang, D., Miller, E. K., Moses, D., Bazan, G. C. & Heeger, A. J. (2000) Macromolecules 33, 5153-5158. [Google Scholar]
- 14.Kabanov A. V., Felgner, P. & Seymour, L. W., (1998) Self-Assembling Complexes for Gene Delivery: From Laboratory to Clinical Trial (Wiley, Chichester, U.K.).
- 15.Kircheis R., Blessing, T., Brunner, S., Wightman, L. & Wagner, E. (2001) J. Controlled Release 7, 165-170. [DOI] [PubMed] [Google Scholar]
- 16.Wolfert M. A., Dash, P. R., Navarova, O., Oupicky, D., Seymour, L. W., Smart, S., Strohalm, J. & Ulbrich, K. M. A. (1999) Bioconjugate Chem. 10, 993-1004. [DOI] [PubMed] [Google Scholar]
- 17.Ganachaud F., Elaïssari, A., Pichot, C., Laayoun, A. & Cros, P. (1997) Langmuir 13, 701-707. [Google Scholar]
- 18.Harada A. & Kataoka, K. (1999) Science 283, 65-67. [DOI] [PubMed] [Google Scholar]
- 19.Bronich T. K., Nguyen, H. K., Eisenberg, A. & Kabanov, A. V. (2000) J. Am. Chem. Soc. 12, 8339-8343. [Google Scholar]
- 20.Nielsen P. E. & Egholm, M., (1999) Peptide Nucleic Acids: Protocols and Applications (Horizon Scientific Press, Portland), pp. 1–19.
- 21.Stender H., Fiandaca, M., Hyldig-Nielsen, J. J. & Coull, J. (2002) J. Microbiol. Methods 48, 1-17. [DOI] [PubMed] [Google Scholar]
- 22.Egholm M., Buchardt, O., Christensen, L., Behrens, C., Freier, S. M., Driver, D. A., Berg, R. H., Kim, S. K., Norden, B. & Nielsen, P. E. (1993) Nature (London) 365, 566-568. [DOI] [PubMed] [Google Scholar]
- 23.Nielsen P. E. (1999) Curr. Opin. Biotechnol. 10, 71-75. [DOI] [PubMed] [Google Scholar]
- 24.Demidov V. V., Potaman, V. N., Frankkamenetskii, M. D., Egholm, M., Buchard, O., Sonnichsen, S. H. & Nielsen, P. E. (1994) Biochem. Pharmacol. 48, 1310-1313. [DOI] [PubMed] [Google Scholar]
- 25.Lakowicz J. R., (1999) Principles of Fluorescence Spectroscopy (Kluwer, New York), pp. 367–394.
- 26.Stork M. S., Gaylord, B. S., Heeger, A. J. & Bazan, G. C. (2002) Adv. Mater. 14, 361-366. [Google Scholar]
- 27.Giesen U., Dleider, W., Berding, C., Geiger, A., Ørum, H. & Nielsen, P. E. (1998) Nucleic Acids Res. 26, 5004-5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomac S., Sarkar, M., Ratilainen, T., Wittung, P., Nielsen, P. E., Nordén, B. & Gräslund, A. (1996) J. Am. Chem. Soc. 118, 5544-5552. [Google Scholar]
- 29.Smith J. O., Olson, D. A. & Armitage, B. A. (1999) J. Am. Chem. Soc. 121, 2686-2695. [Google Scholar]
- 30.Gaylord B. S., Wang, S., Heeger, A. J. & Bazan, G. C. (2001) J. Am. Chem. Soc. 123, 6417-6418. [DOI] [PubMed] [Google Scholar]
- 31.Egholm M., Nielsen, P. E., Berg, R. H., Buchardt, O. & Berg, R. H. (1992) J. Am. Chem. Soc. 144, 9677-9678. [Google Scholar]
- 32.Betts L., Josey, J. A., Veal, J. M. & Jordan, S. R. (1995) Science 270, 1838-1841. [DOI] [PubMed] [Google Scholar]
- 33.Hanvey J. C., Peffer, N. J., Bisi, J. E., Thomson, S. A., Cadilla, R., Josey, J. A., Ricca, D. J., Hassman, C. F., Bonham, M. A., Au, K. G., et al. (1992) Science 258, 1481-1485. [DOI] [PubMed] [Google Scholar]