Abstract
The cellular alterations associated with skeletal muscle differentiation share a high degree of similarity with key phenotypic changes usually ascribed to apoptosis. For example, actin fiber disassembly/reorganization is a conserved feature of both apoptosis and differentiating myoblasts and the conserved muscle contractile protein, myosin light chain kinase, is required for the apoptotic feature of membrane blebbing. As such, these observations suggest that the induction of differentiation and apoptosis in the myogenic lineage may use overlapping cellular mechanisms. Here, we report that skeletal muscle differentiation depends on the activity of the key apoptotic protease, caspase 3. Peptide inhibition of caspase 3 activity or homologous deletion of caspase 3 leads to dramatic reduction in both myotube/myofiber formation and expression of muscle-specific proteins. Subsequently, we have identified Mammalian Sterile Twenty-like kinase as a crucial caspase 3 effector in this cellular process. Mammalian Sterile Twenty-like kinase is cleavage-activated by caspase 3, and restoration of this truncated kinase in caspase 3 null myoblasts restores the differentiation phenotype. Taken together, these results confirm a unique and unanticipated role for a caspase 3-mediated signal cascade in the promotion of myogenesis.
Skeletal myogenesis has served as an ideal model system in which to explore the basic precepts of regulatory control governing cellular differentiation. The understanding of muscle-cell commitment and differentiation has been rapidly advanced with the discovery of a group of transcription factor proteins referred to as the myogenic regulatory factors (MRFs). The primary MRFs, Myf5 and MyoD, are required for the formation, propagation, and survival of skeletal myoblasts. The secondary MRFs, myogenin and MRF4, act later in the program likely as differentiation factors (1). In addition, MRF proteins require secondary interactions with other muscle transcription factors to attain maximal transcriptional activity—i.e., members of the myocyte enhancer factor-2 (MEF2) family. MEF2 belong to a distinct class of factors referred to as MADS domain proteins. MEF2 proteins bind to a cognate site (A,T rich) which is found in the promoters of many muscle-specific genes (2), including both myogenin and MRF4 (3–6). The direct physical interaction of MEF2 proteins and MRFs synergistically enhance transcription (4). As such, a paradigm has emerged suggesting that sequential expression of transcription factor families leads to the initiation and progression of differentiation (2).
The constitutive expression of MRF/MEF2 proteins, even during myoblast growth, strongly suggests that the enhanced activity of these proteins and ultimately the progression of myogenesis depends on posttranslational modifications. Recent evidence demonstrates that select signal transduction pathways may provide the impetus to initiate muscle differentiation and a promyogenic role has been assigned to the mitogen-activated protein kinase (MAPK) pathways. For example, the p38 subfamily of MAPKs play critical roles in the activation of muscle gene transcription. Activation of p38α is concurrent with the induction of differentiation, and use of pharmacological inhibitors of p38α can effectively block this process (7–10). Furthermore, activation of p38α leads to enhanced activity for both MyoD and MEF2A/C; the elevated MEF2 transcriptional activity being a result of p38α directed phosphorylation (11, 12). A promyogenic effect has also been attributed to p38γ, although the mechanism by which this kinase facilitates myogenesis remains to be identified (13).
These observations have implicated a stimulatory role for signal cascade effector proteins in skeletal muscle differentiation. Nevertheless, the precipitating event(s) that leads to an up-regulation of these effector kinases remains unknown. The p38 MAPK family has also been implicated in the initiation and progression of apoptosis (4, 15). One interpretation that could be drawn from these seemingly disparate activation patterns is that apoptosis and differentiation use the same signal cascade to engage MAPK activity. Indeed, such a contention bears significant merit when one considers that differentiating myoblasts share a remarkable similarity with many key cellular alterations endemic to apoptosis. For example, actin fiber disassembly/reorganization is a conserved feature of both apoptosis (16, 17) and differentiating myoblasts (18–20). Second, the conserved muscle contractile protein, myosin light chain kinase, is required for the apoptotic feature of membrane blebbing (21). Finally, increased activity of matrix metalloproteinases seems to be an indispensable requirement for orchestrating membrane fusion in both myoblast differentiation and apoptosis (22, 23). Taken together, these observations suggest that the induction of differentiation and apoptosis in the myogenic lineage may use overlapping cellular mechanisms.
Successful completion of the apoptotic program depends on the activity of a unique class of proteolytic enzymes referred to as caspases (24). Although caspase proteins primarily target and inactivate proteins through serine-directed cleavage events, various caspase proteins also engage apoptosis through cleavage activation of signaling molecules including MEKK1 (25) and SLK (18). In addition, caspase 3 activity has been linked with activation of the MAPKs JNK and p38, albeit through activation of intervening kinases (25–27). Therefore, on the basis of the demonstrated signal efficacy of the death effector protease family, we examined the role of caspase 3-mediated signaling in skeletal muscle differentiation.
Here, we report that early initiation of the skeletal myogenic program relies on the activity of the key apoptotic serine protease, caspase 3. Inhibition of caspase 3 activity and homologous deletion of caspase 3 severely attenuates myoblast fusion and myotube formation. These effects are coincident with reduced myogenic protein accumulation. Subsequently, we have identified a primary caspase 3 target, Mammalian Sterile Twenty-like kinase (MST1), as a crucial effector kinase that mediates caspase 3 activation to effect myogenesis. These results imply that previously conceived proapoptotic death effectors also have decisive context-dependent roles as promyogenic proteins.
Materials and Methods
Myoblast Culture.
C2C12 cultures were maintained as described (3). Caspase activity was measured by using a Caspase 3 and Caspase 8 Fluorometric Assay Kit (PharMingen). Cells in which caspase activity was perturbed were treated with 20 μM inhibitors specific for either caspase 3 (z-DEVD.fmk) or caspase 8 (z-LETD.fmk) (Enzyme Systems Products, Livermore, CA) (based on concentration curve tests of 0–100 μM). Control cells were treated with an equivalent amount of vehicle (DMSO). Primary myoblast cells were derived from skeletal muscles of the hind- and fore-limbs from 1- to 2-day-old C57BL/6 wild-type, caspase 3 heterozygous and caspase 3 null mice (28, 29) (n = 5 for each genotype) as described (30).
Transfection Assays.
The complete ORF of the human MST1 (Stratagene) was inserted into the expression vector pcDNA3.1 Myc/His (Invitrogen). A constitutively active MST1 (MST1-act) was prepared as described (31). Transfections were performed using Lipofectamine Plus (GIBCO).
Gain-of-function experiments were performed in which 20 pg (based on trials of 1–50 pg) of activated caspase 3 protein (Gene Therapy Systems, San Diego) was transfected into a subconfluent population of C2C12 myoblasts by using the BioPorter protein delivery reagent (Gene Therapy Systems).
Protein Isolation, Immunoblot Analyses, and Immunocytochemistry.
Cells from C2C12 and primary cultures were washed with ice-cold PBS and lysed on ice for 45 min in modified radioimmunoprecipitation assay buffer containing 20 mM NaF, 5 mM Na3VO4, and 10 μg/ml each of aprotinin, leupeptin, pepstatin, and PMSF. Immunoblot analyses were performed as described (32) by using antibodies for MST1, and p38α (Upstate Biotechnology, Lake Placid, NY), MEF2C (Cell Signaling Technology, Beverly, MA), cyclin D1, Troponin-T, and MyoD (Santa Cruz Biotechnology), myogenin F5D and myosin heavy chain MF20 mAb (Development Studies Hybridoma Bank, Iowa City), tubulin (Sigma), and active caspase 3 and poly(ADP-ribose) polymerase (PARP) (Biovision, Mountain View, CA).
C2C12 myoblasts were fixed and stained with anti-MF20 as described (30). Active caspase 3 was detected by using a polyclonal active caspase 3 antibody (Biovision). The level of cellular proliferation was assessed by using a monoclonal cyclin D1 antibody (PharMingen). Secondary IgG conjugated to fluorescein (FITC) or rhodamine (Chemicon) or Alexa 594 (Molecular Probes) were used for immunofluorescent labeling.
Coimmunoprecipitation and Kinase Analyses.
Immunoprecipitation was performed with 150–200 μg of total protein, incubated in modified radioimmunoprecipitation assay buffer at 4°C for 16 h containing the respective antibody (32). Immunoprecipitates used for kinase analyses were washed in modified radioimmunoprecipitation assay buffer and incubated in kinase buffer supplemented with [γ-32P]ATP and either recombinant myelin basic protein (MBP) or recombinant Histone H1 (Upstate Biotechnology) as the target substrate. Following electrophoresis, gels were dried and phospho-incorporation was analyzed by autoradiography.
Samples were subjected to in-gel kinase analyses as described (33), with MBP or Histone H1 as the target substrate. For two-dimensional in-gel kinase analyses, samples were isoelectric focused in the first dimension by using pH 4.0–7.0 Immobilized pH Gradient strips (Bio-Rad). Samples were electrophoresed in the second dimension by using SDS-10% polyacrylamide gel containing Histone H1 as the in-gel substrate. Additional in-gel kinase analyses were performed as above and as described (33).
In Vitro Translation and MST1 Proteolysis.
MST1- pcDNA3.1 Myc/His was in vitro transcribed and translated by using a TnT T7 Coupled Wheat Germ Extract System (Promega). MST1 protein labeled with [35S]methionine was incubated for 30 min at 37°C with samples of C2C12 cells after z-DEVD.fmk and low-serum media treatment.
Results and Discussion
Caspase 3−/− Myoblasts Display a Differentiation Deficit.
From our observations and as reported (28, 29), caspase 3 null mice that survive to early perinatal life were strikingly smaller relative to their wild-type and heterozygote littermates. Furthermore, a visible reduction in total skeletal muscle mass was observed (data not shown). As such, we reasoned that myogenesis may be compromised in these animals. To examine the effect of caspase 3 ablation on myogenesis, primary myoblast cultures were generated from 1- to 2-day-old mice carrying a targeted null mutation in caspase 3. The proliferation of wild-type, caspase 3+/−, and caspase 3−/− myoblasts were similar. On induction of differentiation, a severe lack of myotube formation was evident in caspase 3−/− cultures relative to wild-type cells (Fig. 1A; compare a fusion index of 10% vs. 80%, null to wild-type, respectively). In addition, the expression of several differentiation-specific proteins was substantially reduced in caspase 3−/− myoblasts relative to wild-type myoblasts, which included hypophosphorylated MyoD (Fig. 1D), myogenin (Fig. 1E), and Troponin-T (Fig. 1G). Myogenin was ≈2-fold higher in wild-type cells relative to caspase 3 null cells after 12 h of low serum treatment. Additional treatment in low serum resulted in ≈1-fold greater levels of myogenin in wild-type myoblasts. Troponin-T protein did not accumulate as quickly in caspase 3 null myoblasts as in wild-type but reached comparable levels after 2 days of low-serum treatment. Furthermore, in caspase 3−/− myoblasts, the levels of cyclin D1, a marker of cellular proliferation remained elevated compared with wild-type myoblasts (Fig. 1F). These observations suggest that elevated caspase 3 activity is necessary for effective differentiation during the early stages of skeletal myogenesis.
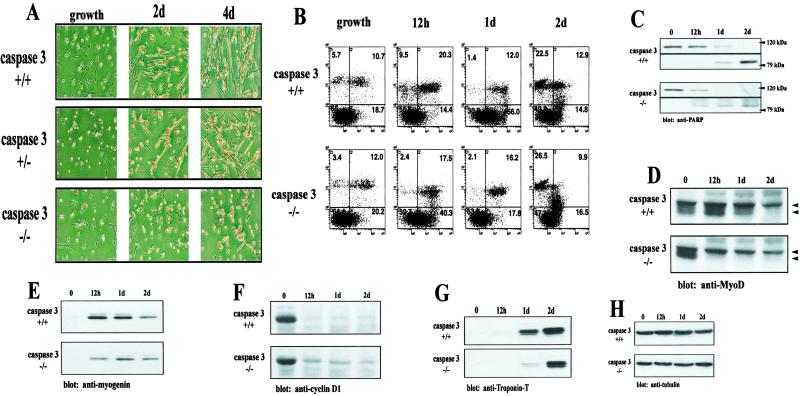
Fig 1.
Caspase 3 null myoblasts have deficient myotube formation. (A) Primary myoblast cultures were derived from wild-type and caspase 3−/− mice (n = 5 for each genotype). Cultures were fixed and stained for myosin heavy chain and counterstained with the nuclear marker hematoxylin (dark staining pattern). A severe lack of myogenesis is noted in caspase 3−/− cells. (B) Flow cytometry analysis of wild-type and caspase 3−/− myoblasts after incubation in low-serum medium. Cells were either left unstained (lower left quadrant), stained with Annexin V-FITC (lower right quadrant), propidium iodide (upper left quadrant), or both (upper right quadrant). A comparable proportion of apoptotic cells was observed for both wild-type and caspase 3 null myoblasts after 2 days of low-serum treatment. (C) PARP cleavage is equivalent in both wild-type and caspase 3 null myoblasts. Both the 118-kDa PARP protein and the 85-kDa cleavage fragment were detected. A reduction of muscle differentiation-specific proteins in caspase 3 null myoblasts was observed by using Western blot analyses for hypophosphorylated MyoD (D) (top and bottom arrows indicate the position of phosphorylated and hypophosphorylated MyoD, respectively), myogenin (E), and Troponin-T (G). The amount of myogenin protein was ≈2-fold higher in wild-type cells after 12 h. Comparable levels of Troponin-T were found after 2 days of low-serum treatment. (F) Caspase 3 null myoblasts displayed a prolonged accumulation of cyclin D1 in the presence of low-serum (differentiation) media. (H) Equal loading for Western blot analyses was assessed by using an anti-tubulin antibody. Band intensity was assessed by using SCION IMAGE.
Conceivably, caspase 3 activity may be required to remove a population of myoblasts (through apoptosis) which would normally inhibit the differentiation process—i.e., a non-cell autonomous effect. Alternatively, elevated caspase 3 activity may lead to intracellular alterations, which in turn activate a differentiation program—i.e., a cell autonomous effect. Therefore, the degrees of apoptosis in caspase null and wild-type myoblasts were compared during low-serum induction of differentiation. Surprisingly, no measurable difference existed in early- or late-stage markers of apoptosis as demonstrated by flow cytometry with Annexin V and propidium iodide staining and analysis of PARP cleavage (Fig. 1 B and C). Furthermore, myocytes from caspase 3 heterozygote mice displayed a reduced level of myogenesis relative to wild-type controls but differentiated nonetheless (Fig. 1A). These results suggest the elevated caspase 3 activity normally associated with differentiation is not strictly apoptotic.
Caspase Activity Has a Positive Role During Skeletal Muscle Differentiation.
The phenomena observed in caspase 3 null animals were further examined in an established myoblast cell population. Because studies have reported changes in the activities of caspases 3 and 8 in skeletal muscle (34–36), we measured the activities of caspases 3 and 8 in the C2C12 skeletal muscle cell line. Proliferating C2C12 cells were placed in low-serum media and allowed to differentiate for 5 days. At the respective time points, the cells were washed extensively to ensure that lysates were free of nonadherent apoptotic cells. Fluorometric analyses revealed a sharp, 9-fold increase in caspase 3 activity within 24 h, which then declined as differentiation progressed (Fig. 2A). Although a similar trend in activity was noted for caspase 8, the increase in activity relative to normal values was not as large as that observed with caspase 3. Immunocytochemical detection of active caspase 3 was used to confirm that the measured increase in caspase 3 activity was a feature of differentiating myoblasts and not a result of cells undergoing apoptosis (data not shown).
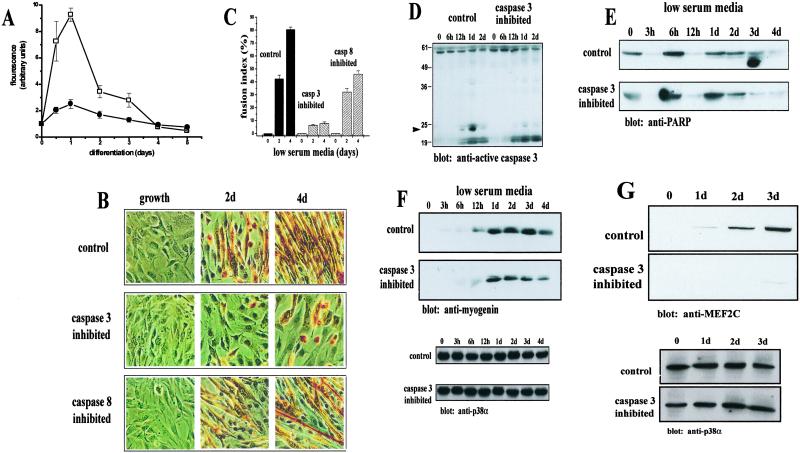
Fig 2.
Caspase 3 activity is required for skeletal muscle differentiation. (A) A sharp increase in caspase 3 activity (□) was found in C2C12 myoblasts after 1 day of low-serum treatment. A much smaller increase in caspase 8 activity (•) was noted, although a similar trend in activation was found (mean ± SEM., n = 5). (B) Morphology of C2C12 cells after inhibition of caspase 3 or caspase 8 activity. Cells were incubated in low-serum media with inhibitors specific for either caspase 3 (Z-DEVD.fmk) or caspase 8 (Z-LETD.fmk) or DMSO (control) and stained with MF20. Myotube formation is drastically attenuated in caspase 3-inhibited cells after a 4-day treatment. A minor attenuation of myotube formation is observed with caspase 8 inhibition. (C) Caspase 3-inhibited cells lack fusion capacity. Calculation of myoblast fusion indices as a percentage of cells containing two or more nuclei within a differentiated myotube (MF20 positive cell). Values were determined as mean ± SEM from three to four independently derived cultures. (D) Western blot analysis of active caspase 3 from control and caspase 3-inhibited cells. The arrow indicates a 21-kDa caspase 3 fragment, indicative of caspase 3 activation abundant in control cells and at very low levels in caspase 3-inhibited samples. (E) Comparable levels of PARP cleavage in both control and caspase 3-inhibited cells. PARP cleavage was accessed by Western blot. Shown is the 118-kDa PARP species. (F) Accumulation of myogenin is delayed and is in lower abundance in caspase 3-inhibited C2C12 myoblasts. The level of myogenin in control cells is approximately twice that of in caspase 3-inhibited cells by day 4. Equal loading was accessed by Western blot with anti-p38α (shown below). (G) Caspase 3-inhibited C2C12 myoblasts express very low levels of MEF2C relative to control cells. Equal loading (shown below) was accessed as in F. Band intensity was assessed by using SCION IMAGE.
As a compliment to our analyses of the caspase 3 null model, we directly targeted caspase activity in C2C12 myoblasts by using pharmacological inhibitors specific for caspase 3 (z-DEVD.fmk) and 8 (z-LETD.fmk). Caspase 3-inhibited myoblasts displayed an impaired formation of myotubes (Fig. 2 B Middle and C). Specifically, these cells remained mononucleated and unfused after 4 days of low-serum conditions relative to vehicle (DMSO)-treated C2C12 myoblasts. Western analyses with a caspase 3-specific antibody on peptide-treated and control cell lysates confirmed that z-DEVD.fmk treatment was effective in inhibiting caspase 3 activity (Fig. 2D). The lack of myoblast differentiation with z-DEVD.fmk-treated C2C12 cells was remarkably similar to caspase 3−/− myoblasts. In contrast, differentiation was more evident in cells that were incubated with caspase 8 inhibitor, z-LETD.fmk (Fig. 2 B Bottom and C). However, the degree of differentiation remained smaller than that observed in vehicle-treated cells. Although a proportion of caspase 3-inhibited cells were able to express the muscle transcription factor myogenin (Fig. 2F), a delayed and attenuated accumulation was evident when compared with control myoblasts. The level of myogenin in control cells consistently remained ≈1.5–2.5 fold higher than in caspase 3-inhibited cells as determined through densitometric quantitation (not shown). More strikingly, the expression of the late myogenic marker MEF2C was virtually absent in cells inhibited for caspase 3 activity (Fig. 2G). Clearly, the deficit in myogenesis brought about by caspase 3 inhibition suggests that caspase 3 is a promyogenic protease and that its effects are greater than the effects mediated by caspase 8. In addition, PARP cleavage products accumulated to a similar degree in z-DEVD-treated and untreated C2C12 myoblasts (Fig. 2E), suggesting that caspase 3 activity is not responsible for the ambient rate of apoptosis of differentiating myoblast populations.
To further test a cell autonomous role of caspase 3 in inducing myoblast differentiation, active caspase 3 protein was transfected into a subconfluent population of C2C12 myoblasts maintained in growth media. Transfected myoblasts initiated the differentiation program, as measured by immunodetection of myosin heavy chain (Fig. 3A; transfection efficiency ≈10%; transfected cells undergoing differentiation >95%, n = 4 independent transfections). Myoblasts expressing myosin heavy chain were also positive for the transfected active caspase 3 protein (not shown) and did not display any morphologic features usually attributed to apoptosis. Moreover, the transfection of active caspase 3 was permissive to inducing myoblast differentiation irrespective of growth-stimulating conditions—i.e., high-serum conditions (Fig. 3A).
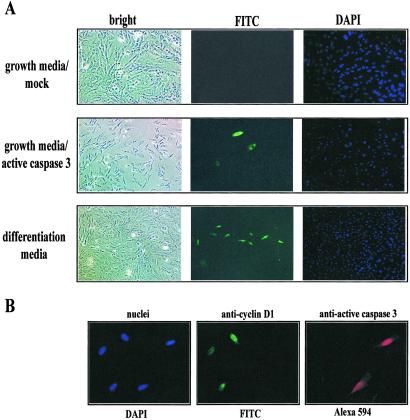
Fig 3.
Activated caspase 3 induces differentiation in growing myoblasts. (A) Subconfluent C2C12 myoblasts were transfected with 20 pg of active caspase 3 protein and examined after 12 h for the presence of myosin heavy chain (anti-MF20) detected by using a FITC-conjugated secondary antibody. A confluent population of cells were incubated in low-serum media (differentiation media) to induce myoblast differentiation and examined after 12 h. In all cells examined, nuclei were localized with DAPI staining. Figure is representative of seven trials. (B) Caspase 3 activity results in a withdrawal of cells from the cell cycle. A subconfluent population of cells maintained in growth media was transfected with 20 pg of active caspase 3 protein. Cells were fixed after 24–28 h and stained for the cell proliferation marker cyclin D1 (FITC-labeled) and active caspase 3 (Alexa 594). Cells containing active caspase 3 do not express cyclin D1. Total nuclei are shown stained with DAPI. Figure is representative of five trials.
Additional experiments using this gain-of-function approach revealed that cells transfected with activated caspase 3 protein did not express the cellular proliferation marker, cyclin D1 (Fig. 3B). Furthermore, cells that stained positive for active caspase 3 were negative for the cell cycle markers phospho-histone H1/H3 (not shown). Therefore, these cells likely exited the cell cycle and began terminal differentiation. Taken together, our observations from both the animal and cell culture models support the hypothesis that endogenous caspase 3 activity is required for myoblast differentiation.
Caspase 3 Activates MST1 Kinase During Differentiation.
When processed to an active form, caspase 3 is capable of targeting several protein kinases and rendering them active, usually through cleavage and removal of their regulatory C-terminal domain (25, 37). Therefore, caspase 3 activation may be necessary to provoke a kinase(s) that is part of the myogenic signaling cascade. Indeed, various promyogenic kinases from the MAPK family (8, 9, 14) have been shown to be cleavage-activated by caspase 3 or associated with upstream kinase effectors that are themselves stimulated through a caspase-directed cleavage event (25, 37). We used a two-dimensional in-gel kinase assay with MBP as a target substrate to identify protein kinases that may be targeted by caspase 3 activity. After a 12-h low-serum treatment of C2C12 myoblasts with z-DEVD.fmk, many endogenous kinases had a diminished level of activity when compared with untreated myoblasts (Fig. 4A). Similar results were seen by using histone H1 as the in-gel substrate (data not shown).
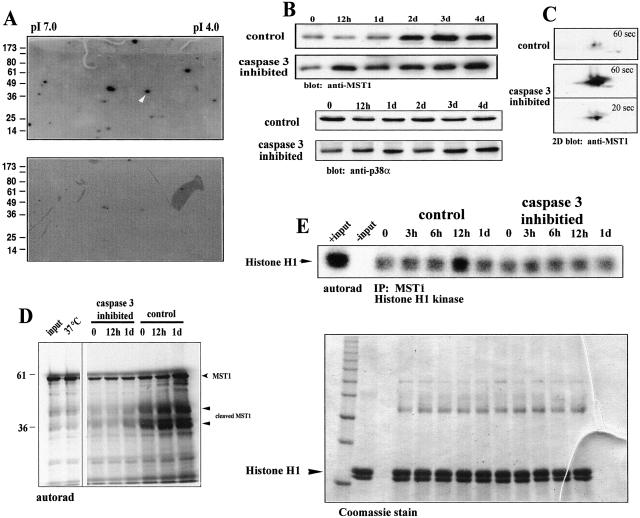
Fig 4.
MST1 is activated by caspase 3 during skeletal muscle differentiation. (A) Two-dimensional in-gel kinase assay. Caspase 3 inhibition results in a significant loss of MBP-directed cellular kinase activities. The putative location of active MST1 kinase, based on predicted pI (5.2) and molecular weight (36 kDa), is indicated by the white arrowhead. (B) Western blot analysis of C2C12 cells by using an anti-MST1 antibody. Control-treated cells show a varied amount of MST1 during 0 and 1 day of differentiation and exemplify a proteolytic-associated event. The level of MST1 protein does not change on the addition of low-serum (differentiation) media in caspase 3-inhibited myoblasts. Shown below is loading control by using p38α. (C) Two-dimensional isoelectric focusing/Western blot of control and caspase 3-treated cells after 12 h in low-serum media. Subsequent immunoblot analysis was performed by using an anti-MST1 antibody. Shown are 1-min exposures for both control and caspase 3-inhibited samples. A 20-s exposure for the caspase 3 sample is included as a comparison of signal intensity. (D) [35S]methionine shows that labeling MST1 is cleaved in differentiating myoblasts. The right-hand side shows the position of [35S]Met-MST1. The arrows below indicate MST1 cleavage products of approximately 36 and ≈45 kDa. (E) MST1 kinase activity increases during myoblast differentiation. [γ-32P]ATP-labeled Histone H1 after immunoprecipitation of MST1 from C2C12 myoblasts under differentiation conditions. A substantial activation of MST1 in the control sample after 12 h of differentiation coincides with the cleavage of MST1 as shown in A and C. Sample loading was indexed by staining the gel with Coomassie blue before drying and autoradiography (E Bottom). The arrow indicates the position of histone H1.
One kinase that was altered in activity comigrated with a predicted pI and molecular weight consistent with MST1. MST1 is a known caspase 3-activated kinase homologous to the yeast family of serine/threonine Ste20 kinases (14, 31). MST1 has been described to act as an upstream kinase effector targeting MKK6 and members of the p38 MAPK family (14, 39). Analyses by Western blot revealed a lower level of full-length MST1 during the initial 24 h of low-serum treatment compared with z-DEVD.fmk-treated cultures (Fig. 4B). In contrast, MST1 protein remained relatively unchanged in z-DEVD.fmk-treated cultures. This difference was further exemplified by using two-dimensional immunoblot analyses (Fig. 4C). To verify that the difference in MST1 accumulation was caused by caspase 3-mediated cleavage, we incubated [35S]methionine-labeled MST1 with protein lysates isolated from both control and z-DEVD.fmk-treated C2C12 cultures. The full-length MST1, migrated to approximately 58 kDa on an SDS-10% polyacrylamide gel. When incubated with lysates from normally differentiating C2C12 cells, a lower band of approximately 36 kDa appeared, indicative of a cleaved MST1 fragment (Fig. 4D). An additional band of approximately 45 kDa was also present in these samples. Recent reports have identified two sites within the MST1 amino acid sequence that are susceptible to caspase cleavage (39). One of these sites, between amino acids 323 and 327, is the conserved caspase 3 cleavage motif DEVD. Cleavage at this site generates a 36-kDa catalytically active fragment (14). Cleavage at a second site (TMTD) between amino acids 346 and 349 generates a 41-kDa fragment, which may represent a functional caspase 8 cleavage domain. In C2C12 cells, the lower 36-kDa MST1 fragment was not apparent in samples where caspase 3 activity was inhibited suggesting that MST1 is usually cleaved by caspase 3 during the early phase of myogenesis. Given that the targeted cleavage of MST1 is tightly coupled to its activation, we examined the activity of MST1 during early periods of differentiation. MST1 was immunoprecipitated from control and z-DEVD.fmk-treated lysates and incubated with Histone H1 in an in vitro kinase assay. Histone H1 was strongly phosphorylated in control lysates after 12 h in low-serum media (>4-fold increase), whereas no phosphorylation at greater than basal levels was detectable from z-DEVD.fmk-treated cells (Fig. 4E). Together, these results support a mechanism whereby caspase 3 mediates the activation of MST1.
Earlier reports have implicated members of the p38 MAPK pathway as substrates for MST1 (14, 39). These kinases have been demonstrated to promote myogenesis by phosphorylating and increasing the endogenous activity of skeletal muscle transcription factors (10, 13). Therefore, we examined their potential as downstream substrates in a caspase 3/MST1 signal conduit. Caspase 3 inhibition led to a block in the activation for both MKK6 and p38γ (data not shown). Taken together, these data indicate that MST1 serves to enhance downstream members of the MAPK cascade that effectively promote skeletal muscle differentiation.
Active MST1 Rescues Myogenesis in Caspase 3−/− Myoblasts.
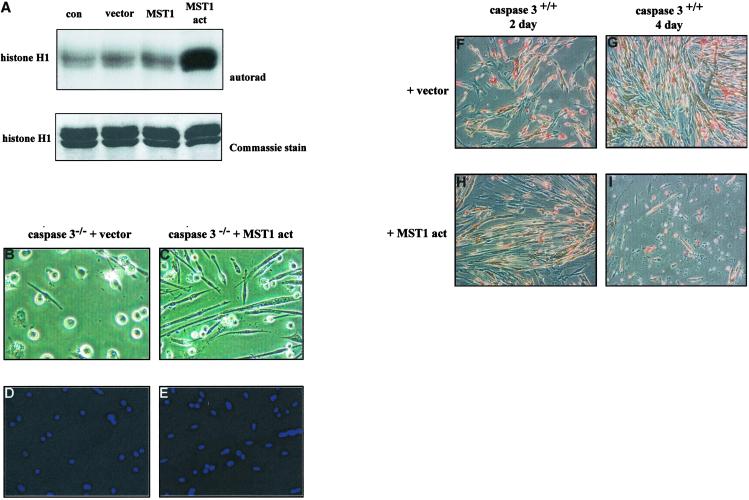
To delineate the effect of MST1 on myogenesis, we transiently transfected either plasmid alone, wild-type MST1, or an activated MST1 (amino acids 1–330, MST1-act) into primary myoblasts derived from caspase 3−/− mice. The activity of these constructs was verified by histone H1-targeted kinase analysis of MST1 from transiently transfected COS1 cells (Fig. 5A). Caspase 3−/− myoblasts transfected with either vector or wild-type MST1 remained as mononucleated myoblasts (Fig. 5 B and D). In contrast, caspase 3−/− myoblasts transfected with MST1-act had elongated and fused with neighboring cells to form multinucleated myotubes (Fig. 5 C and E). Therefore, the expression of activated MST1 in caspase 3−/− myoblasts rescued the aberrant myogenic phenotype. To demonstrate further the promyogenic role of activated MST1, we transfected MST1-act into wild-type primary myoblasts. Transfected wild-type cultures left in differentiation media for 2 days had phenotypic characteristics that were comparable to wild-type cultures after 4 days of differentiation (compare Fig. 5 H and G). Furthermore, we observed that MST1-act-transfected cells subject to a protracted differentiation time course (>4 days) displayed enhanced apoptosis (Fig. 5I). These results suggest that the endogenous activation pattern of MST1 accelerates skeletal myocyte differentiation, although potent apoptotic events can be engaged by this kinase during extended periods of activation.
Fig 5.
Activated MST1 rescues the caspase 3−/− myoblast phenotype. (A) Kinase activity of MST1 protein constructs. MST1-act-transfected cells display a high level of kinase activity. Accuracy of sample loading is indicated by the Coomassie-stained gel shown below (n = 4). (B) Caspase 3 knockout myoblasts transfected with vector alone failed to differentiate. (C) Caspase 3 knockout myoblasts containing MST1-act differentiated to form fused, multinucleated myotubes. The efficiency of differentiation in MST1-act myoblasts was >85%. Total nuclei were visualized by using DAPI staining (D and E). (F–I) Wild-type myoblasts were transfected with vector or with MST1-act and allowed to differentiate for up to 4 days. After 2 days of differentiation, cells containing MST1-act (H) share a similar phenotype with vector-transfected cultures differentiated for 4 days (G). Extensive cell death is observed when cells are transfected with MST1-act and differentiated for 4 days (I).
We have observed that both biologic and chemical blockade of caspase 3 results in a profound deficiency in myogenesis. We demonstrate that the Ste20-like kinase MST1, serves as a fundamental conduit in mediating caspase 3 induction of muscle differentiation. Previous reports have suggested that caspase 3 is capable of regulating nonapoptotic functions in certain cell types—i.e., nuclear extrusion in differentiating lens epithelium and keratinocytes (40, 41), and T cell activation (42). Nevertheless, our observations directly link caspase 3 activity to such profound alterations in cell morphology without ushering in a death-like phenotype. Of interest, similar signaling components (MST1, MKK6, p38) are also found in numerous differentiated cell types including hepatocytes, thymocytes, and neurons (43–45). In view of these observations, it is reasonable to suggest that caspase 3/MST1 signaling may be an indispensable component for initiating cellular differentiation in general. As such, any substantial differentiation deficit may account for a considerable proportion of the excessive neuronal cell numbers found in caspase 3 null mutants (28, 45). Although such a broad hypothesis has not been directly examined, the current study provides a critical rationale for such an investigation.
Acknowledgments
We thank M. Rudnicki, D. Picketts, and S. Kolodziejczyk for helpful discussions, as well as Tammy-Lynn Tremblay, Emma Tibo, and Karl Parato for their most excellent assistance. This work was supported by grants to L.A.M. from the Canadian Institutes of Health Research (CIHR) and the Muscular Dystrophy Association, and by a grant to R.S.S. from CIHR. L.A.M. and R.S.S. are CIHR scholars. P.F. is supported by a Postdoctoral Fellowship from the Heart and Stroke Foundation of Canada.
Abbreviations
MRF, myogenic regulatory factor
MEF, myocyte enhancer factor
MAPK, mitogen-activated protein kinase
MST1, Mammalian Sterile Twenty-like kinase
MBP, myelin basic protein
PARP, poly(ADP-ribose) polymerase
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Megeney L. A. & Rudnicki, M. A. (1995) Biochem. Cell. Biol. 73, 723-732. [DOI] [PubMed] [Google Scholar]
- 2.Black B. L. & Olson, E. N. (1998) Annu. Rev. Cell Dev. Biol. 14, 167-196. [DOI] [PubMed] [Google Scholar]
- 3.Black B. L., Martin, J. F. & Olson, E. N. (1995) J. Biol. Chem. 270, 2889-2892. [DOI] [PubMed] [Google Scholar]
- 4.Molkentin J. D., Black, B. L., Martin, J. F. & Olson, E. N. (1995) Cell 83, 1125-1136. [DOI] [PubMed] [Google Scholar]
- 5.Yee S. P. & Rigby, P. W. (1993) Genes Dev. 7, 1277-1289. [DOI] [PubMed] [Google Scholar]
- 6.Cheng T. C., Wallace, M. C., Merlie, J. P. & Olson, E. N. (1993) Science 261, 215-218. [DOI] [PubMed] [Google Scholar]
- 7.Cuenda A. & Cohen, P. (1999) J. Biol. Chem. 274, 4341-4346. [DOI] [PubMed] [Google Scholar]
- 8.Wu Z., Woodring, P. J., Bhakta, K. S., Tamura, K., Wen, F., Fermisco, J. R., Karin, M., Wang, J. Y. J. & Puri, P. L. (2000) Mol. Cell. Biol. 20, 3951-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zetser A., Greding, E. & Bengal, E. (1999) J. Biol. Chem. 274, 5193-5200. [DOI] [PubMed] [Google Scholar]
- 10.Ornatsky O. I., Cox, D. M., Tangirala, P., Andreucci, J. J., Quinn, Z. A., Wrana, J. L., Prywes, R., Yu, Y. T. & McDermott, J. C. (1999) Nucleic Acids Res. 27, 2646-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang S. H., Galanis, A. & Sharrocks, A. D. (1999) Mol. Cell. Biol. 19, 4028-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puri P. L., Wu, Z., Zhang, P., Wood, L. D., Bhakta, K. S., Han, J., Fermisco, J. R., Karin, M. & Wang, J. Y. J. (2000) Genes Dev. 14, 574-584. [PMC free article] [PubMed] [Google Scholar]
- 13.Lechner C., Zahalka, M. A., Giot, J.-F., Moller, N. P. & Ullrich, A. (1996) Proc. Natl. Acad. Sci. USA 93, 4355-4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graves J. D., Gotoh, Y., Draves, K. E., Ambrose, D., Han, D. K., Wright, M., Chernoff, J., Clark, E. A. & Krebs, E. G. (1998) EMBO J. 17, 2224-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Juo P., Kuo, C. J., Reynolds, S. E., Konz, R. F., Raingeaud, J., Davis, R. J., Biemann, H. P. & Blenis, J. (1997) Mol. Cell. Biol. 17, 24-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall A. & Nobes, C. D. (2000) Philos. Trans. R. Soc. London B 355, 965-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coleman M. L. & Olson, M. F. (2002) Cell Death Differ. 5, 493-504. [DOI] [PubMed] [Google Scholar]
- 18.Sabourin L. A. & Rudnicki, M. A. (2000) Clin. Genet. 57, 16-25. [DOI] [PubMed] [Google Scholar]
- 19.Gallo R., Serafini, M., Castellani, L., Falcone, G. & Alema, S. (1999) Mol. Biol. Cell 10, 3137-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qu G., Yan, H. & Strauch, A. R. (1997) J. Cell. Biochem. 67, 514-527. [PubMed] [Google Scholar]
- 21.Mills J. C., Stone, N. L., Erhardt, J. & Pittman, R. N. (1998) J. Cell Biol. 140, 627-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Powell W. C., Fingleton, B., Wilson, C. L., Boothby, M. & Matrisian, L. M. (1999) Curr. Biol. 9, 1441-1447. [DOI] [PubMed] [Google Scholar]
- 23.Yagami-Hiromasa T., Sato, T., Kurisaki, T., Kamijo, K., Nabeshima, Y. & Fujisawa-Sehara, A. (1995) Nature (London) 377, 652-656. [DOI] [PubMed] [Google Scholar]
- 24.Utz P. J. & Anderson, P. (2000) Cell Death Differ. 7, 589-602. [DOI] [PubMed] [Google Scholar]
- 25.Cardone M. H., Salvesen, G. S., Widmann, C., Johnson, G. & Frisch, S. M. (1997) Cell 90, 315-323. [DOI] [PubMed] [Google Scholar]
- 26.Chaudhary P. M., Eby, M. T., Jasmin, A. & Hood, L. (1999) J. Biol. Chem. 274, 19211-19219. [DOI] [PubMed] [Google Scholar]
- 27.Frasch S. C., Nick, J. A., Fadok, V. A., Bratton, D. L., Worthen, G. S. & Henson, P. M. (1998) J. Biol. Chem. 273, 8389-8397. [DOI] [PubMed] [Google Scholar]
- 28.Kuida K., Zheng, T. S., Na, S., Kuan, C., Yang, D., Karasuyama, H., Rakic, P. & Flavell, R. A. (1996) Nature (London) 384, 368-372. [DOI] [PubMed] [Google Scholar]
- 29.Cregan S. P., MacLaurin, J. G., Craig, C. G., Roberston, G. S., Nicholson, D. W., Park, D. S. & Slack, R. S. (1999) J. Neurosci. 19, 7860-7869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Megeney L. A., Kablar, B., Garrett, K., Anderson, J. E. & Rudnicki, M. A. (1996) Genes Dev. 10, 1173-1183. [DOI] [PubMed] [Google Scholar]
- 31.Creasy C. L. & Chernoff, J. (1995) J. Biol. Chem. 270, 21695-21700. [DOI] [PubMed] [Google Scholar]
- 32.Kolodziejczyk S. M., Wang, L., Balazsi, K., DeRepentigny, Y., Kothary, R. & Megeney, L. A. (1999) Curr. Biol. 9, 1203-1206. [DOI] [PubMed] [Google Scholar]
- 33.Kameshita I. & Fujisawa, H. (1989) Anal. Biochem. 183, 139-143. [DOI] [PubMed] [Google Scholar]
- 34.Belizario J. E., Lorite, M. J. & Tisdale, M. J. (2001) Br. J. Cancer 84, 1135-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yaoita Y. & Nakajima, K. (1997) J. Biol. Chem. 272, 5122-5127. [DOI] [PubMed] [Google Scholar]
- 36.Mukasa T., Momoi, T. & Momoi, M. Y. (1999) Biochem. Biophys. Res. Commun. 260, 139-142. [DOI] [PubMed] [Google Scholar]
- 37.Widmann C., Gerwins, P., Johnson, N. L., Jarpe, M. B. & Johnson, G. L. (1998) Mol. Cell. Biol. 18, 2416-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun S. & Ravid, K. (1999) J. Cell. Biochem. 76, 44-60. [DOI] [PubMed] [Google Scholar]
- 39.Graves J. D., Draves, K. E., Gotoh, Y., Krebs, E. G. & Clark, E. A. (2001) J. Biol. Chem. 276, 14909-14915. [DOI] [PubMed] [Google Scholar]
- 40.Ishizaki Y, Jacobson, M. D. & Raff, M. C. (1998) J. Cell. Biol. 140, 153-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weil M., Raff, M. C. & Braga, V. M. (1999) Curr. Biol. 9, 361-364. [DOI] [PubMed] [Google Scholar]
- 42.Alam A., Cohen, L. Y., Aouad, S. & Sekaly, R. P. (1999) J. Exp. Med. 190, 1879-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng T. S., Schlosser, S. F., Dao, T., Hingorani, R., Crispe, I. N., Boyer, J. L. & Flavell, R. A. (1998) Proc. Natl. Acad. Sci. USA 95, 13618-13623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ko H. W., Han, K. S., Kim, E. Y., Ryu, B. R., Yoon, W. J., Jung, Y. K., Kim, S. U. & Gwag, B. J. (2000) J. Neurochem. 74, 2455-2461. [DOI] [PubMed] [Google Scholar]
- 45.Woo M., Hakem, R., Soengas, M. S., Duncan, G. S., Shahinian, A., Kagi, D., Hakem, A., McCurrach, M., Khoo, W., Kaufman, S. A., et al. (1998) Genes Dev. 12, 806-819. [DOI] [PMC free article] [PubMed] [Google Scholar]