Abstract
The CC-chemokine receptor 5 (CCR5) is the major coreceptor for the entry of macrophage-tropic (R5) HIV-1 strains into target cells. Posttranslational sulfation of tyrosine residues in the N-terminal tail of CCR5 is critical for high affinity interaction of the receptor with the HIV-1 envelope glycoprotein gp120 in complex with CD4. Here, we focused on defining precisely the sulfation pattern of the N terminus of CCR5 by using recombinant human tyrosylprotein sulfotransferases TPST-1 and TPST-2 to modify a synthetic peptide that corresponds to amino acids 2–18 of the receptor (CCR5 2–18). Analysis of the reaction products was made with a combination of reversed-phase HPLC, proteolytic cleavage, and matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry (MALDI-TOF MS). We found that CCR5 2–18 is sulfated by both TPST isoenzymes leading to a final product with four sulfotyrosine residues. Sulfates were added stepwise to the peptide producing specific intermediates with one, two, or three sulfotyrosines. The pattern of sulfation in these intermediates suggests that Tyr-14 and Tyr-15 are sulfated first, followed by Tyr-10, and finally Tyr-3. These results represent a detailed analysis of the multiple sulfation reaction of a peptide substrate by TPSTs and provide a structural basis for understanding the role of tyrosine sulfation of CCR5 in HIV-1 coreceptor and chemokine receptor function.
The CC-chemokine receptor 5 (CCR5) is a member of the protein superfamily of G protein-coupled receptors (GPCRs) (1–3). High-affinity binding of the CC-chemokines MIP-1α, MIP-1β, RANTES (1–3), or MCP-2 (4) to CCR5 induces signaling through G proteins of the Gi subfamily (1) and leads to chemotactic responses in CCR5-expressing leukocytes (5).
In addition to their physiological function in chemokine signaling, some chemokine receptors are used as coreceptors by HIV-1. Entry of HIV-1 into target cells is mediated by the sequential interaction of the envelope glycoprotein gp120 with CD4 and a chemokine receptor on the cell membrane (6). CCR5 and CXCR4 are the primary HIV-1 coreceptors in vivo (7, 8). CCR5, in particular, is the principal coreceptor for macrophage-tropic HIV-1 strains (R5 isolates) that are commonly transmitted between individuals (6, 9). A naturally occurring CCR5 mutant (Δ32) with a deletion in the second extracellular loop results in impaired membrane expression of the receptor and leads to resistance to HIV-1 infection in homozygous individuals (6).
Interaction with both types of CCR5 ligands, CC-chemokines and the HIV-1 gp120-CD4 complex, involves the N-terminal domain, as well as other extracellular regions of the receptor (10–13). Within the N-terminal domain, a region rich in tyrosine residues and acidic amino acid residues (Fig. 1; residues 2–18) was identified as a major determinant of HIV-1 coreceptor function (11, 12, 14–17). Sequence similarities with proteins known to be modified by tyrosine O-sulfation, a posttranslational modification mediated by tyrosylprotein sulfotransferases (TPSTs) in the trans-Golgi network (18), led to the recent discovery of tyrosine sulfation within this region of CCR5 (19).
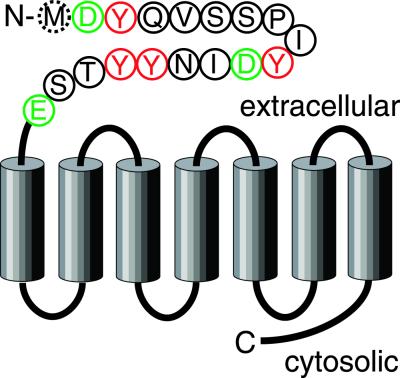
Fig 1.
Schematic representation of human CCR5 with potential tyrosine sulfation sites. The seven transmembrane helices are shown as cylinders. Connecting extracellular and cytosolic loops as well as N- and C-terminal domains are depicted as black lines. Amino acid residues in the N-terminal sequence corresponding to peptide CCR5 2–18 are represented in single-letter codes. Met-1, which is not present in CCR5 2–18, is marked by a dashed circle. The potentially sulfated tyrosine residues at positions 3, 10, 14, and 15 are highlighted in red, and the acidic amino acid residues Asp-2, Asp-11, and Glu-18 are shown in green.
Although the role of tyrosine sulfation in protein function is not generally well understood, in the case of CCR5 it has been shown that tyrosine sulfation is crucial for efficient gp120–CD4 binding and HIV-1 coreceptor function (19–21). Sulfotyrosines were also reported to contribute to the binding of the CC-chemokines MIP-1α and MIP-1β to CCR5 (19, 21).
The specific contribution of the four potential tyrosine sulfation sites in CCR5 to the HIV-1 coreceptor function was investigated in different studies by using either a mutagenesis approach (15, 19) or binding and competition experiments with synthetic sulfotyrosine peptides (20, 21). While these studies agree on the importance of sulfotyrosines at positions 10 and 14, the role of Tyr-3 and Tyr-15 remains in question.
Here we present the pattern observed by enzymatic in vitro sulfation of a peptide corresponding to amino acids 2–18 of CCR5 (CCR5 2–18) by using recombinant human TPSTs. We analyzed intermediates and the final product of the sulfation reaction by using a combination of reversed-phase (RP)-HPLC, proteolytic cleavage, and matrix-assisted laser desorption/ionization–time-of-flight MS (MALDI-TOF MS). By using this in vitro approach, we found that CCR5 2–18 is sulfated by the two known human TPSTs, TPST-1 (22) and TPST-2 (23, 24), resulting in a final product in which all four tyrosine residues are sulfated. We also found that sulfates are added stepwise to the peptide and that Tyr-14 and Tyr-15 are sulfated first, followed by Tyr-10, and finally Tyr-3. These results represent a detailed analysis of the multiple sulfation reaction of a peptide substrate by TPSTs and provide a basis for understanding the role of posttranslational tyrosine sulfation of CCR5 in HIV-1 coreceptor function and chemokine signaling.
Experimental Procedures
Reagents.
Peptides CCR5 2–18 (DYQVSSPIYDINYYTSE-NH2) and PSGL-1 1–15 (amino acids 1–15 of the P-selectin glycoprotein ligand-1, QATEYEYLDYDFLPE-NH2) were synthesized as C-terminal amides by the solid-phase method (25), using Fmoc chemistry (26) and purified by RP-HPLC (The Rockefeller University Protein/DNA Technology Center, New York). Expression vectors pMSH1TH and pMSH2TH encoding recombinant engineered human TPST-1 and TPST-2 were a gift of K. L. Moore (University of Oklahoma Health Sciences Center, Oklahoma City). Anti-protein C resin and sequencing grade proteases endoproteinase Asp-N, chymotrypsin, and carboxypeptidase Y were obtained from Roche Applied Science (Indianapolis). All other reagents were of analytical grade and from Sigma or Fisher Scientific (Pittsburgh).
Expression and Purification of Recombinant Engineered Human TPSTs.
Human TPST-1 and TPST-2 were expressed as soluble variants lacking the cytoplasmic N terminus and the transmembrane domain, which are not required for enzymatic activity. A transferrin signal peptide followed by a protein C epitope was N-terminally fused to the catalytic domain of both TPSTs (23). HEK293-T cells were transfected with plasmids pMSH1TH or pMSH2TH with LipofectAMINE Plus (Invitrogen). After 48 h, plates were washed once with PBS and cells were harvested in ice-cold PBS supplemented with the protease inhibitor mixture Complete (Roche Applied Science, Indianapolis). TPSTs were then partially purified from HEK293-T cell extracts by anti-protein C immunoaffinity chromatography (22).
In Vitro Sulfation of Peptides.
Stock solutions of unsulfated peptides at 10 mg/ml were prepared in DMSO and diluted in sulfation buffer (40 mM Pipes, pH 6.8/300 mM NaCl/20 mM MnCl2/50 mM NaF/1% Triton X-100/1 mM 5′-AMP) (22, 23) to final concentrations of 0.1 mg/ml. TPST-1, TPST-2, or a mixture (1:1) of both enzymes (40 or 180 μg/ml total) and the sulfation cosubstrate 3-phosphoadenosine-5-phosphosulfate (PAPS) (400 μM) were added and the reaction mixtures were incubated at 37°C for PSGL-1 1–15 or 16°C for CCR5 2–18. At selected time points, aliquots of the sulfation reactions were analyzed by RP-HPLC.
RP-HPLC of Sulfotyrosine Peptides.
Samples were analyzed by RP-HPLC, using an analytical C18 column (218TP54, Vydac, Hesperia, CA; 5 μm, 300 Å). Separation was achieved with a linear gradient from 1 to 95% eluent B over 40 min at a flow rate of 1.0 ml/min (eluent A, 20 mM ammonium acetate, pH 6.5, in water; eluent B, 20 mM ammonium acetate, pH 6.5, in 70% acetonitrile). For analytical runs, injection volumes were 20–60 μl. To purify up to 20 μg of sulfotyrosine peptides for further MS and proteolytic analysis, repetitive injections were done. Eluting fractions corresponding to peptide peaks were collected and the solvent was evaporated. Samples for MALDI-TOF MS were redissolved in 150 μl of a 1:2 (vol/vol) mixture of water and acetonitrile and the solvent was evaporated again to remove residual ammonium acetate.
Proteolytic Cleavage of Sulfotyrosine Peptides.
Purified sulfotyrosine peptides (4–20 μg) were dissolved in 5 μl of DMSO plus 60 μl of 20 mM Tris⋅HCl, pH 7.0. Endoproteinase Asp-N (120 ng) was added and the mixtures were incubated for 4 h at 37°C. N-terminal fragments created by endoproteinase Asp-N (CCR5 2–10) and purified by RP-HPLC were further cleaved with chymotrypsin. The enzyme (2.5 μg) was added to 0.1 to 2 μg of the fragments in 25 μl of 100 mM Tris⋅⋅HCl, pH 7.8/10 mM CaCl2 and the mixtures were incubated for 24 h at 25°C. For carboxypeptidase Y cleavage, RP-HPLC-purified sulfotyrosine peptides (2 μg) were dissolved in 10 μl of 50 mM sodium citrate, pH 6.0. Carboxypeptidase Y (300 ng) was added and the mixtures were incubated for 2–4 h at 25°C. After separation by RP-HPLC, cleavage products were identified by MALDI-TOF MS.
MALDI-TOF MS.
Sulfotyrosine peptide samples were analyzed by using α-cyano-4-hydroxycinnamic acid (Sigma) as a matrix. Solvent conditions were optimized to prevent disruption of labile thioester bonds. The matrix was prepared as a saturated solution in a 2:1 (vol/vol) mixture of water and acetonitrile. Samples were diluted 1:10 in matrix solution and a small aliquot (0.5 μl) of peptide-matrix solution was spotted onto the sample plate by using an ultra-thin layer method (27). All mass measurements were performed on a Voyager DE-STR (Applied Biosystems, Foster City, CA) MALDI-TOF MS, operating in negative linear, delayed extraction mode. Spectra from 200 individual laser shots were averaged (2 ns data channel width), calibrated externally and internally, and further analyzed using the program M-over-Z (http://www.proteometrics.com).
Results
Enzymatic in Vitro Sulfation of CCR5 2–18.
We used the PSGL-1 1–15 peptide to test the sulfation activity of expressed enzymes TPST-1 and TPST-2 (22, 23). The concentration of the sulfation cosubstrate PAPS (400 μM) was adjusted to allow for the stoichiometric sulfation of the peptide. By using RP-HPLC, we found effective incorporation of sulfates into PSGL-1 1–15 (data not shown).
In initial experiments with CCR5 2–18, we used a 1:1 mixture of TPST-1 and TPST-2 to optimize reaction conditions. A time course of CCR5 2–18 sulfation was carried out at different reaction temperatures and the reaction products were detected by RP-HPLC. We found that sulfation was most effective at 16°C and progressed for at least 100 h (Fig. 2A). These conditions gave reproducibly high sulfate incorporation into the CCR5 2–18 peptide and were used throughout the following experiments.
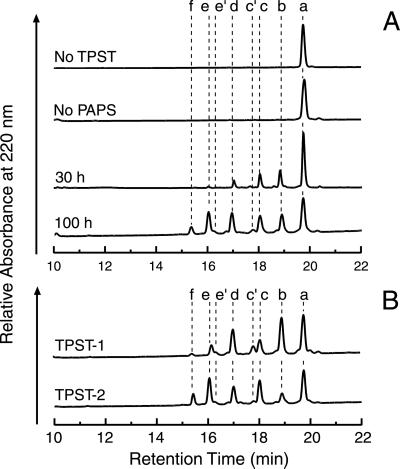
Fig 2.
RP-HPLC analysis of CCR5 2–18 sulfation products. (A) Characterization of the in vitro sulfation reaction. Peptide CCR5 2–18 (0.1 mg/ml, ≈50 μM) was incubated with a mixture of TPST-1 and TPST-2 (20 μg/ml each) and in the presence of the sulfation cosubstrate PAPS (400 μM). After 30 h or 100 h at 16°C, 60 μl aliquots were analyzed by RP-HPLC. In negative-control experiments (100 h incubation time), either the TPST mixture (no TPST) or PAPS (no PAPS) was omitted. Peaks were labeled a–f in increasing order of hydrophilicity. (B) Comparison of TSPT-1 and TPST-2. CCR5 2–18 (0.1 mg/ml, ≈50 μM) was incubated for 100 h with TPST-1 (40 μg/ml) or TPST-2 (40 μg/ml) in the presence of PAPS (400 μM).
As shown in Fig. 2A, the reaction required the presence of cosubstrate PAPS and TPST enzymes, indicating specific TPST-catalyzed sulfation. After 100 h, the reaction produced six major peaks which were labeled a–f. In addition, two minor peaks, labeled c′ and e′, were produced. The most hydrophobic peak, a, elutes at a time similar to the unreacted peptide. Peaks b–f appeared in a time-dependent fashion, with retention times decreasing in increments of about 0.8 min. The formation of several sulfation products with different hydrophilicities suggests that these represent CCR5 2–18 species that differ in the number and positions of tyrosine sulfates.
The peptide was then incubated for 100 h with TPST-1 or TPST-2 individually. As shown in Fig. 2B, both TPST-1 and TPST-2 are able to modify CCR5 2–18. Although the two isoenzymes produced similar peak patterns, there were distinct differences in the relative intensities of individual peaks (Fig. 2B). For the sake of simplicity, the methodology used to count sulfates and localize sulfation sites will be illustrated in detail only for major peaks a–f prepared with the TPST mixture. The same methodology was applied for sulfated species found with TPST-1 or TPST-2 individually.
MALDI-TOF MS of Sulfation Products.
Peptide species separated by RP-HPLC were analyzed by MALDI-TOF MS to determine the exact number of sulfates present in each species. Peak fraction a was first analyzed in positive ion reflector mode (Mr = 2,055.2 Da) and confirmed to be CCR5 2–18 (theoretical Mr = 2,054.9 Da). However, the positive ion mode is inappropriate for the analysis of sulfated species because of the addition of negative charges to an already acidic peptide and the lability of the sulfoester bond (28). Thus, fractions were analyzed in linear negative ion mode. Masses were determined with a precision exceeding 500 ppm, allowing for the unambiguous identification of each MS peak. As seen in Fig. 3, fraction a produced a peak corresponding to nonsulfated CCR5 2–18 with a small sodium adduct peak, whereas fractions b–f showed series of peaks of increasing complexity. This complexity arises from the superimposition of two series of ion peaks, one corresponding to loss of sulfate (ΔMr = −80 Da) to various degrees, the other corresponding to a distribution of sodium adducts. Fraction f, for example, contains CCR5 2–18 peptide with four sulfates, which appears in the spectrum as 3-, 4-, or 5-sodium adduct peaks. However, the spectrum is dominated by peaks with lower sulfate numbers, because of fragmentation during mass spectrometric analysis. Each of these loss-of-sulfate species is itself accompanied by satellite sodium adduct peaks.
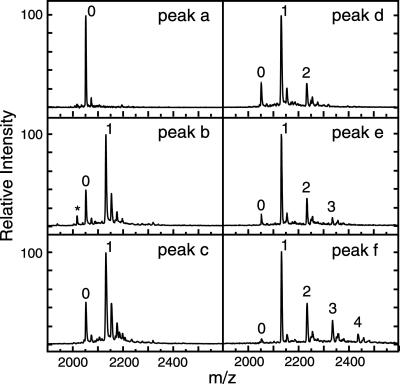
Fig 3.
MALDI-TOF MS of CCR5 2–18 sulfation products. Peak fractions a–f from RP-HPLC were analyzed in negative ion mode by using an ultra-thin layer sample preparation method (27). Because of the lability of tyrosine sulfo–ester bonds under MS conditions (28), spectra for highly sulfated peptide species were dominated by loss-of-sulfate ions. Multisulfated peptide species could only be detected as sodium adducts. Peaks were labeled according to the number of identified sulfates as shown in Table 1. Ions with additional sodium adducts were not labeled. In peak fraction b, a small amount of the ion [(CCR5 3–18 + SO3) − H]− (*) was detected, consistent with the loss of the initial Asp.
Although the sulfated peptide ions displayed a high degree of instability, the relatively higher stability of their sodium adducts allowed us to determine the number of sulfates present in each HLPC-purified peptide. These numbers are reported for each peak fraction in Fig. 3 and Table 1. Taken together, the MALDI-TOF MS data show that the products of the sulfation reaction represent CCR5 2–18 species with zero (peak a), one (peaks b and c), two (peak d), three (peak e), or four (peak f) sulfotyrosines.
Table 1.
MALDI-TOF MS of sulfation products
| Peak | MS label | Ion | No. of sulfates |
|---|---|---|---|
| a | 0 | [(CCR5 2–18) − H]− | None |
| b | 1 | [(CCR5 2–18 + SO3) − H]− | 1 |
| c | 1 | [(CCR5 2–18 + SO3) − H]− | 1 |
| d | 2 | [(CCR5 2–18 + 2SO3) + Na − 2H]− | 2 |
| e | 3 | [(CCR5 2–18 + 3SO3) + 2Na − 3H]− | 3 |
| f | 4 | [(CCR5 2–18 + 4SO3) + 3Na − 4H]− | 4 |
Proteolytic Cleavage of CCR5 2–18 Sulfation Products.
To localize modified tyrosines within the various sulfation products of CCR5 2–18, we generated fragments containing subsets of the four tyrosines by proteolysis with endoproteinase Asp-N, chymotrypsin, and carboxypeptidase Y (Fig. 4A). All unambiguously identified sulfation sites are reported in Table 2, Major components.
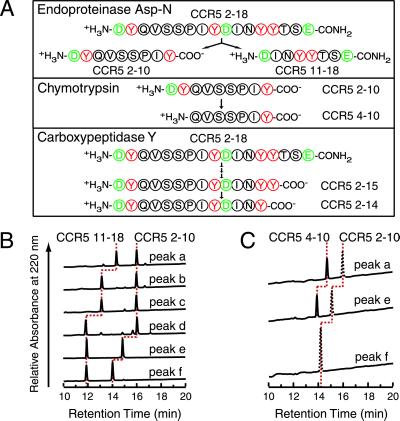
Fig 4.
Analysis of sulfation sites in CCR5 2–18 sulfation products. (A) Strategy for the generation of proteolytic fragments. Fragments CCR5 2–10 and CCR5 11–18 were produced by endoproteinase Asp-N cleavage. Fragment CCR5 4–10 was generated from fragment CCR5 2–10 by chymotrypsin cleavage. For clarity, the dipeptide fragment (DY), which was not further analyzed, is not shown. Fragments CCR5 2–15 and CCR5 2–14 were obtained by carboxypeptidase Y cleavage of CCR5 2–18 sulfation products. (B) RP-HPLC chromatograms of endoproteinase Asp-N cleavage products. Nonsulfated CCR5 2–18 (peak a) and tyrosine sulfation products (peaks b–f), cleaved with endoproteinase Asp-N, were analyzed by RP-HPLC and identified by MALDI-TOF MS as fragments CCR5 2–10 and CCR5 11–18 containing zero, one, or two sulfates (MS data not shown). In the RP-HPLC chromatograms, peaks corresponding to the same fragment are connected by dotted red lines. The stepwise decrease in the elution time for a given fragment indicates incorporation of one or two sulfates. (C) RP-HPLC chromatograms of chymotrypsin cleavage products. Asp-N-generated fragments CCR5 2–10 were further cleaved with chymotrypsin. Peaks identified as uncleaved starting material CCR5 2–10 are drawn with dotted black lines, whereas peaks corresponding to the cleavage product CCR5 4–10 are drawn with solid black lines. Peaks corresponding to the same peptide sequence are connected by dotted red lines.
Table 2.
Localization of sulfotyrosines by analysis of cleavage fragments
| Peak | Uncleaved | Endoproteinase Asp-N | Chymotrypsin | Carboxypeptidase Y | Sulfotyrosines | TPST | ||
|---|---|---|---|---|---|---|---|---|
| Major components | ||||||||
| a | 2–18 | 2–10 | 11–18 | 4–10 | n.d. | n.d. | none | 1, 2 |
| b | 2–18(SO3) | 2–10 | 11–18(SO3) | n.d. | 2–15(SO3) | 2–14(SO3) | 14 | 1, 2 |
| c | 2–18(SO3) | 2–10 | 11–18(SO3) | n.d. | 2–15(SO3) | 2–14 | 15 | 1, 2 |
| d | 2–18(SO3)2 | 2–10 | 11–18(SO3)2 | n.d. | n.d. | n.d. | 14, 15 | 1, 2 |
| e | 2–18(SO3)3 | 2–10(SO3) | 11–18(SO3)2 | 4–10(SO3) | n.d. | n.d. | 10, 14, 15 | 1, 2 |
| f | 2–18(SO3)4 | 2–10(SO3)2 | 11–18(SO3)2 | Not cleavable | n.d. | n.d. | 3, 10, 14, 15 | 1, 2 |
| Minor components | ||||||||
| b | 2–18(SO3) | 2–10(SO3) | 11–18 | Not cleavable | n.d. | n.d. | 3 | 2 |
| c | 2–18(SO3)2 | 2–10(SO3) | 11–18(SO3) | Not cleavable | n.d. | n.d. | 3, 14 | 1 |
| c′ | 2–18(SO3)2 | 2–10(SO3) | 11–18(SO3) | 4–10(SO3) | n.d. | n.d. | 10, 14 | 1 |
| d | 2–18(SO3)2 | 2–10(SO3) | 11–18(SO3) | 4–10(SO3) | n.d. | n.d. | 10, 15 | 2 |
| e′ | 2–18(SO3)2 | 2–10(SO3) | 11–18(SO3)2 | Not cleavable | n.d. | n.d. | 3, 14, 15 | 1 |
n.d., not determined.
Inhibition of chymotrypsin cleavage by sulfotyrosines at position 3.
Distinction between position 14 and 15 only on the basis of RP-HPLC retention times.
In the case of TPST-2 amounts were found insufficient for analysis.
RP-HPLC peak fractions a–f (Fig. 2) were cleaved with endoproteinase Asp-N, which cleaves CCR5 2–18 at a single site between Tyr-10 and Asp-11 (Fig. 4A). RP-HPLC chromatograms of the cleavage products (Fig. 4B) show two major peaks of similar intensity for all fractions. MALDI-TOF MS identified the first HPLC peak as CCR5 11–18 and the second as CCR5 2–10. The number of sulfotyrosines within each fragment was determined by MALDI-TOF MS (data not shown). According to this data, b and c both harbor a single sulfate within the C-terminal half of the molecule, which would indicate that one is sulfated at Tyr-14 and the other at Tyr-15. The main component in peak d contains two sulfates in the C-terminal part, which shows that Tyr-14 and Tyr-15 are both sulfated (Table 2, Major components). Peak e contains two sulfates in the C-terminal part and a single sulfate in the N-terminal part, which means that it is sulfated either at Tyr-10 or Tyr-3, in addition to Tyr-14 and Tyr-15. Peak f, as expected for a CCR5 2–18 species with four sulfates, has two sulfates in each of the C- and N-terminal halves.
To determine which of the two tyrosine residues in the N-terminal half of e was modified, we cleaved this fragment with chymotrypsin (Fig. 4A). For comparison we also analyzed the corresponding fragments from a and f. RP-HPLC analysis (Fig. 4C), followed by MALDI-TOF MS of the purified chymotrypsin products showed that the N-terminal fragment of a and e gave rise to the CCR5 4–10 fragment. This fragment was sulfated when the starting material was e, and, as expected, unsulfated for a. Peak f, on the other hand, was resistant to chymotrypsin cleavage. These results show that e is sulfated at Tyr-10 and suggest that sulfation at Tyr-3 in f inhibits cleavage by chymotrypsin. This in turn provides confirmation that Tyr-3 in e is not sulfated (Table 2, Major components).
To determine which of the residues Tyr-14 or Tyr-15 was sulfated in the C-terminal half of b and c, we cleaved these sulfopeptides with carboxypeptidase Y, which removes amino acids stepwise from the C terminus of the peptide (Fig. 4A). We found that peak b is sulfated at Tyr-14, whereas peak c is sulfated at Tyr-15 (Table 2, Major components).
Based on the observed sulfation species and time dependent studies (not shown) we postulate that the sulfation reaction progresses through the observed intermediates in a sequential manner (Scheme 1).
 |
Scheme 1.
Sulfation Pattern with Individual TPSTs.
A complete analysis was also performed for the major sulfation products generated by TPST-1 and TPST-2 separately. The same sulfation sites were identified for each product as for the enzyme mixture (Table 2, Major components), which indicates that the path described in Scheme 1 is also valid for the individual enzymes.
To further substantiate the sequential nature of the sulfation reaction and examine the differences between TPST-1 and TPST-2, we analyzed the time course of CCR5 2–18 sulfation for each enzyme. CCR5 2–18 was incubated with either TPST-1 or TPST-2 in the presence of PAPS, and the products analyzed at different time points by using RP-HPLC. The time-dependent buildup and depletion of sulfation products is shown for TPST-1 in Fig. 5A and for TPST-2 in Fig. 5B. Because the reaction is performed with an excess of peptide substrate over enzyme, the observed kinetics and amounts of intermediates show that the substrate has to be released after each sulfation step.
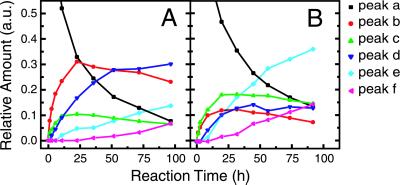
Fig 5.
Time course of CCR5 2–18 sulfation catalyzed by TPST-1 and TPST-2. CCR5 2–18 (0.1 mg/ml) was incubated with TPST-1 (A) or TPST-2 (B) (180 μg/ml each) in the presence of PAPS (400 μM) at 16°C. At the indicated time points, 40-μl aliquots were analyzed by RP-HPLC, and relative amounts for the different peptide species (a–f) were calculated from the peak areas.
In the course of the reaction with each enzyme, unsulfated CCR5 2–18 (peak a) was depleted and the different sulfation products appeared sequentially. Peaks b and c were formed first and in parallel. Only after a significant amount of b and c had accumulated was d observed. Similarly, e was formed after d accumulated, and finally f was formed as the end product of the sulfation reaction. Although the reaction sequences seem to be similar for TPST-1 and TPST-2, there are significant differences in the rates of formation and relative quantities of the different products. For example, b was formed at a higher level than c by TPST-1, whereas the opposite was observed for TPST-2. Similarly, the relative quantities of d and e depended on the enzyme used. Most strikingly, the final product f was formed at a lower rate by TPST-1 than by TPST-2. However, the overall rates of sulfate incorporation into CCR5 2–18 were similar for TPST-1 and TPST-2.
The reaction with either enzyme did not lead to full sulfation of all four tyrosines. This could be explained by a progressive decline in sulfation activity in our conditions caused by cosubstrate degradation, a loss of enzyme activity, or product inhibition. Consequently, no clear-cut conclusion can be drawn regarding the completeness of the sulfation reaction.
Analysis of Side Products Created by TPST-1 Versus TPST-2.
In the course of analyzing the major products generated by the individual TPSTs, we also characterized less abundant side products. Two of these products, labeled c′and e′, were resolved by RP-HPLC (Fig. 2B), whereas others were only resolved after cleavage and further chromatographic separation. Using the same combination of RP-HPLC, MALDI-TOF MS, and proteolytic cleavage as described for the major products, we also localized minor sulfation sites. As reported in Table 2, Minor components, some differences appear in the side products generated by the two enzymes.
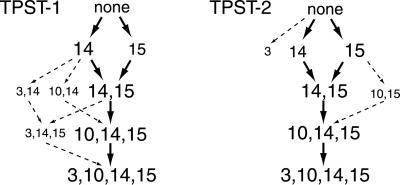
By comparing the intensities of all intermediates in RP-HPLC, we evaluated the relative preponderance of intermediates for both TPSTs. The paths of sulfation for TPST-1 and TPST-2 are presented along with their respective variations in Fig. 6.
Fig 6.
Reaction scheme for the sequential sulfation of CCR5 2–18 by TPST-1 and TPST-2. CCR5 2–18 species that arise from the sulfation of nonsulfated CCR5 2–18 (none), with either TPST-1 (Left) or TPST-2 (Right), are represented by the sulfotyrosine positions. Major sulfation products are depicted as bold numbers and corresponding reaction pathways are represented by bold arrows. Minor sulfation products are in smaller fonts and the corresponding reaction pathways are depicted as dotted arrows.
Discussion
It is well established that the sulfation of tyrosine residues in the N terminus of CCR5 is required for optimal HIV-1 coreceptor and chemokine receptor function of CCR5 (19–21). However, it has not been clear which of the four tyrosines are sulfated in the mature wild-type receptor, and whether there is a site preference for sulfation. The substrate specificity of the two known TPSTs has not been determined, and CCR5 sensitivity to sulfation in general cannot be predicted for lack of a well defined consensus sequence. Site-directed mutagenesis alone cannot address these difficult questions unequivocally because amino acid replacements can affect sulfation at neighboring sites (19). Our approach to examine sulfation by using a reconstituted in vitro system yielded useful data and provided information that should prove valuable for future study of the receptor itself.
CCR5 2–18 was found to be effectively sulfated by the two known human tyrosylprotein sulfotransferases, TPST-1 (22) and TPST-2 (23, 24). With both isoenzymes, we observed the incorporation of multiple sulfates (up to four) into the peptide in a nonrandom, sequential fashion. Interestingly, only 5 of 15 possible sulfated peptide species were predominant. Based on the time course of the reaction and the structures of the sulfation intermediates, a mechanism can be proposed in which the tyrosines at positions 14 or 15 are sulfated first, followed by 10, and then 3 (Fig. 6).
Studies addressing the functional significance of specific sulfotyrosines in the N terminus of CCR5 have mainly focused on the HIV-1 coreceptor function. The importance of sulfotyrosines at positions 10 and 14 has been demonstrated in several studies by using either CCR5 mutants (19) or partially sulfated peptides (20, 21). The contribution of positions 3 and 15, on the other hand, is less well understood (15, 19, 20). Our in vitro results for CCR5 2–18 show that position 10 is sulfated after positions 14 and 15, suggesting that a CCR5 species with sulfotyrosines at positions 10 and 14, but not at 15, is less likely to exist. Tyr-3 was reported to be involved in coreceptor function (15), but sulfation of this residue had no effect in one study (20), and only a weak effect in another study (19). The discrepancies between the different functional studies could, in part, originate from the use of different HIV-1 strains, which may depend on different sulfation patterns of CCR5.
A number of studies have focused on defining the sequence requirements for tyrosine sulfation (ref. 29 and references therein). The general conclusion from these studies is that for sulfation to occur, a tyrosine residue needs to be in proximity to one or more acidic residues. Although it seems that most of the information required for sulfation to occur is contained within ±5 residues around the tyrosine, no single consensus sequence has yet been defined. Furthermore, because tyrosine sulfation sites are often clustered, sulfation of one site may depend on the negative charges of sulfates already present at other sites. Interestingly, we observed independent sulfation of two directly adjacent tyrosines, Tyr-14 and Tyr-15. Effective sulfation of a third (Tyr-10) and fourth tyrosine (Tyr-3), on the other hand, occurred after Tyr-14 and Tyr-15 had been sulfated. This apparent order of sulfation events suggests that the sulfation of Tyr-10 and Tyr-3 may depend on the prior sulfation of Tyr-14 and Tyr-15. Alternatively, it may reflect different intrinsic sulfation kinetics at these sites. Most likely, it is the result of a combination of both factors.
The role of tertiary structure elements of CCR5 not present in the N-terminal peptide or components of the cellular sulfation machine presently unknown remains to be determined. Neither is it yet clear whether processing of CCR5 in vivo leads to a homogeneously or heterogeneously sulfated receptor population in the cell membrane. Our in vitro results suggest that if partially sulfated, the receptor population is unlikely to be homogenous.
Sulfation of tyrosine residues is a posttranslational modification found in secreted proteins and in extracellular regions of membrane proteins of multicellular eukaryotes (30, 31). It has been estimated that up to 1% of all tyrosines of the eukaryotic proteome may be sulfated (32). Among the growing number of proteins that actually have been shown to be modified by tyrosine sulfation are molecules that are important for hemostasis, chemotaxis, inflammation, and development, as well as for viral and possibly cancer pathogenesis (31). Although the functional significance of tyrosine sulfation has been investigated in only a few systems, there is evidence that this modification may be important for protein–protein interactions in general (31). Sulfated peptides are potentially useful tools to probe these interactions, as has been shown for the interaction of CCR5 with gp120 (20, 21, 33), and they could turn out to be useful in the development of new drugs. Chemical methods for the synthesis of sulfated peptides exist. However, the sensitivity of the tyrosine sulfoester bond to strong acids tends to result in low overall yields. Peptides with multiple sulfotyrosines are often difficult to obtain. In these cases, enzymatic sulfation may be the method of choice to obtain the desired product.
In summary, this study presents a detailed analysis of the multiple sulfation reaction of a peptide substrate of TPST-1 and TPST-2. The methods described are useful for the preparation of peptides with multiple sulfotyrosines and to investigate potential tyrosine sulfation sites. The results provide a structural basis for understanding the role of posttranslational tyrosine sulfation of CCR5 in HIV-1 cellular entry and in chemokine receptor function, and may provide a basis for the design of therapeutic agents aimed at blocking HIV-1 cellular entry.
Acknowledgments
We thank Dr. K. L. Moore for the gift of TPST expression vectors, and Dr. C. G. Unson for helpful discussions. This work was supported by a grant from the National Institutes of Health (RR00862 from the National Center for Research Resources). C.S. is an Associate and T.P.S. is an Associate Investigator of the Howard Hughes Medical Institute.
Abbreviations
CCR5, CC-chemokine receptor 5
MALDI-TOF MS, matrix-assisted laser desorption/ionization–time-of-flight MS
PAPS, 3′-phosphoadenosine 5′-phosphosulfate
RP, reversed-phase
TPST, tyrosylprotein sulfotransferase
References
- 1.Combadiere C., Ahuja, S. K., Tiffany, H. L. & Murphy, P. M. (1996) J. Leukocyte Biol. 60, 147-152. [DOI] [PubMed] [Google Scholar]
- 2.Samson M., Labbe, O., Mollereau, C., Vassart, G. & Parmentier, M. (1996) Biochemistry 35, 3362-3367. [DOI] [PubMed] [Google Scholar]
- 3.Raport C. J., Gosling, J., Schweickart, V. L., Gray, P. W. & Charo, I. F. (1996) J. Biol. Chem. 271, 17161-17166. [DOI] [PubMed] [Google Scholar]
- 4.Gong W., Howard, O. M., Turpin, J. A., Grimm, M. C., Ueda, H., Gray, P. W., Raport, C. J., Oppenheim, J. J. & Wang, J. M. (1998) J. Biol. Chem. 273, 4289-4292. [DOI] [PubMed] [Google Scholar]
- 5.Murphy P. M., Baggiolini, M., Charo, I. F., Hebert, C. A., Horuk, R., Matsushima, K., Miller, L. H., Oppenheim, J. J. & Power, C. A. (2000) Pharmacol. Rev. 52, 145-176. [PubMed] [Google Scholar]
- 6.Berger E. A., Murphy, P. M. & Farber, J. M. (1999) Annu. Rev. Immunol. 17, 657-700. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y., Lou, B., Lal, R. B., Gettie, A., Marx, P. A. & Moore, J. P. (2000) J. Virol. 74, 6893-6910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y. J. & Moore, J. P. (1999) J. Virol. 73, 3443-3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dragic T. (2001) J. Gen. Virol. 82, 1807-1814. [DOI] [PubMed] [Google Scholar]
- 10.Rucker J., Samson, M., Doranz, B. J., Libert, F., Berson, J. F., Yi, Y., Smyth, R. J., Collman, R. G., Broder, C. C., Vassart, G., et al. (1996) Cell 87, 437-446. [DOI] [PubMed] [Google Scholar]
- 11.Ross T. M., Bieniasz, P. D. & Cullen, B. R. (1998) J. Virol. 72, 1918-1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blanpain C., Doranz, B. J., Vakili, J., Rucker, J., Govaerts, C., Baik, S. S., Lorthioir, O., Migeotte, I., Libert, F., Baleux, F., et al. (1999) J. Biol. Chem. 274, 34719-34727. [DOI] [PubMed] [Google Scholar]
- 13.Howard O. M., Shirakawa, A. K., Turpin, J. A., Maynard, A., Tobin, G. J., Carrington, M., Oppenheim, J. J. & Dean, M. (1999) J. Biol. Chem. 274, 16228-16234. [DOI] [PubMed] [Google Scholar]
- 14.Dragic T., Trkola, A., Lin, S. W., Nagashima, K. A., Kajumo, F., Zhao, L., Olson, W. C., Wu, L., Mackay, C. R., Allaway, G. P., et al. (1998) J. Virol. 72, 279-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rabut G. E., Konner, J. A., Kajumo, F., Moore, J. P. & Dragic, T. (1998) J. Virol. 72, 3464-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farzan M., Choe, H., Vaca, L., Martin, K., Sun, Y., Desjardins, E., Ruffing, N., Wu, L., Wyatt, R., Gerard, N., et al. (1998) J. Virol. 72, 1160-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doranz B. J., Lu, Z. H., Rucker, J., Zhang, T. Y., Sharron, M., Cen, Y. H., Wang, Z. X., Guo, H. H., Du, J. G., Accavitti, M. A., et al. (1997) J. Virol. 71, 6305-6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baeuerle P. A. & Huttner, W. B. (1987) J. Cell Biol. 105, 2655-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farzan M., Mirzabekov, T., Kolchinsky, P., Wyatt, R., Cayabyab, M., Gerard, N. P., Gerard, C., Sodroski, J. & Choe, H. (1999) Cell 96, 667-676. [DOI] [PubMed] [Google Scholar]
- 20.Cormier E. G., Persuh, M., Thompson, D. A., Lin, S. W., Sakmar, T. P., Olson, W. C. & Dragic, T. (2000) Proc. Natl. Acad. Sci. USA 97, 5762-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farzan M., Vasilieva, N., Schnitzler, C. E., Chung, S., Robinson, J., Gerard, N. P., Gerard, C., Choe, H. & Sodroski, J. (2000) J. Biol. Chem. 275, 33516-33521. [DOI] [PubMed] [Google Scholar]
- 22.Ouyang Y. B., Lane, W. S. & Moore, K. L. (1998) Proc. Natl. Acad. Sci. USA 95, 2896-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ouyang Y. B. & Moore, K. L. (1998) J. Biol. Chem. 273, 24770-24774. [DOI] [PubMed] [Google Scholar]
- 24.Beisswanger R., Corbeil, D., Vannier, C., Thiele, C., Dohrmann, U., Kellner, R., Ashman, K., Niehrs, C. & Huttner, W. B. (1998) Proc. Natl. Acad. Sci. USA 95, 11134-11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merrifield R. B. (1963) J. Am. Chem. Soc. 85, 2149-2154. [Google Scholar]
- 26.Carpino L. A. & Han, G. Y. (1972) J. Org. Chem. 37, 3404-3409. [Google Scholar]
- 27.Cadene M. & Chait, B. T. (2000) Anal. Chem. 72, 5655-5658. [DOI] [PubMed] [Google Scholar]
- 28.Gibson B. W. & Cohen, P. (1990) Methods Enzymol. 193, 480-501. [DOI] [PubMed] [Google Scholar]
- 29.Bundgaard J. R., Vuust, J. & Rehfeld, J. F. (1997) J. Biol. Chem. 272, 21700-21705. [DOI] [PubMed] [Google Scholar]
- 30.Huttner W. B. & Baeuerle, P. A. (1988) Mod. Cell Biol. 6, 97-140. [Google Scholar]
- 31.Kehoe J. W. & Bertozzi, C. R. (2000) Chem. Biol. 7, R57-R61. [DOI] [PubMed] [Google Scholar]
- 32.Baeuerle P. A. & Huttner, W. B. (1985) J. Biol. Chem. 260, 6434-6439. [PubMed] [Google Scholar]
- 33.Cormier E. G., Tran, D. N., Yukhayeva, L., Olson, W. C. & Dragic, T. (2001) J. Virol. 75, 5541-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]