Abstract
Five different enzymatic activities, catalyzed by both microsomal and mitochondrial cytochrome P450 monooxygenases (CYPs), are strongly implicated in the biosynthesis of ecdysone (E) from cholesterol. However, none of these enzymes have been characterized completely. The present data show that the wild-type genes of two members of the Halloween family of embryonic lethals, disembodied (dib) and shadow (sad), code for mitochondrial cytochromes P450 that mediate the last two hydroxylation reactions in the ecdysteroidogenic pathway in Drosophila, namely the C22- and C2-hydroxylases. When sad (CYP315A1) is transfected into Drosophila S2 cells, the cells metabolize 2-deoxyecdysone (2dE) to E and the [3H]ketotriol (2,22-dideoxyecdysone) to 22-deoxyecdysone. In contrast, dib (CYP302A1) is responsible for the conversion of the [3H]ketotriol to [3H]2dE. When cells are transfected with both dib and sad, they metabolize the [3H]ketotriol to [3H]E in high yield. The expression of sad and dib is concentrated within the individual segments of the developing epidermis when there is a surge of ecdysteroid midway through embryogenesis. This result occurs before the ring gland has developed and suggests that the embryonic epidermis is a site of ecdysteroid biosynthesis. This pattern then diminishes, and, during late embryogenesis, expression of both genes is concentrated in the prothoracic gland cells of the developing ring gland. Expression of dib and sad continues to be localized in this endocrine compartment during larval development, being maximal in both the late second and third instar larvae, about the time of the premolt peaks in the ecdysteroid titer.
Steroid hormones control many aspects of reproduction, development, and homeostasis in higher organisms (1). In arthropods, steroid hormones play equal or even more vital developmental roles, especially in mediating transitions between developmental stages (2, 3). Indeed, the first evidence that steroid hormones act at the transcriptional level came from studies of the effects of ecdysone (E) on dipteran polytene chromosomes more than four decades ago (2, 3). The molting of larval Drosophila melanogaster, for example, is initiated by the synthesis and release of the steroid hormone E from the prothoracic gland cell portion of the ring gland in response to a neuroendocrine signal (2). E is then converted to the principal molting hormone, 20-hydroxyecdysone (20E), by a P450 enzyme in peripheral target tissues. The regulatory effects of 20E are mediated through activation of a nuclear hormone receptor, resulting in the transcriptional cascades appropriate for eliciting specific developmental events (3, 4).
To facilitate the identification and characterization of the enzymes responsible for ecdysteroid biosynthesis in Drosophila, we used a combined molecular genetic and biochemical approach (5) that was based on recent data that identified the product of the disembodied (dib) locus as being potentially involved in the biosynthesis of ecdysteroids and as a member of a new family of cytochrome P450 monooxygenase (CYP)-type enzymes (i.e., CYP302A1; ref. 6). Mutations in dib result in a number of striking embryonic phenotypes, including disruptions in head involution, dorsal closure, gut morphogenesis, and a failure to produce embryonic cuticle. These developmental disruptions are accompanied by, or are a result of, low titers of ecdysteroids, as is the severe reduction in the epidermal expression of the 20E inducible gene IMP-E1. Several other mutations, including shadow (sad) and collectively termed the Halloween group, exhibit similar phenotypes (6, 7). We reasoned that at least some of these genes might also encode enzymes involved in E biosynthesis, and perhaps code for CYPs.
Materials and Methods
Insects.
The Drosophila mutant strains, dib and sad, were obtained from the Bloomington Stock Center (Bloomington, IN) and raised on standard cornmeal/yeast extract/dextrose medium (6). Brain-ring gland complexes were dissected from late second, early third, and late third instar wild-type Drosophila larvae synchronized at molting, and ovaries were from mature adults. The latter two tissues were also used for subsequent in vitro incubation with various radiolabeled intermediates (8, 9). Prothoracic glands were dissected from late fifth instar Manduca sexta larvae and frozen (−70°C) until thawed and incubated with various radiolabeled intermediates (10–12).
Ecdysteroid Intermediates.
High specific activity 22,23-[3H]-5β[H]-3β,14α,25-trihydroxycholesta-7-ene-6-one ([3H]ketotriol, [3H]2,22-dideoxyecdysone, [3H]2,22dE, 60 Ci/mmol) was a gift from C. Kappler (Université Louis Pasteur, Strasbourg, France). 2-Deoxyecdysone (2dE) was a gift from R. Lafont (Université P. et M. Curie, Paris). E and 20E standards were purchased from Sigma/Aldrich. The [3H]2dE and [3H]22-deoxyecdysone ([3H]22dE) metabolites were produced by the brief incubation of extensively homogenized M. sexta prothoracic glands with the [3H]ketotriol (12). [3H]E (60 Ci/mmol) was purchased from NEN.
Confirmation of sad as CYP315A1.
Adult heterozygous (sad/TM3) genomic Drosophila mutant DNA (two alleles) were amplified by PCR using the primers 5′-AGCGTTTCACAGCCGAGAGC-3′ and 5′-TGCTGCAGGCATTGTTTTCCTAG-3′. Sequencing used the Thermosequenase cycle sequencing kit (United States Biochemical) according to the manufacturer's protocol. In each resulting sequence, both mutant sad alleles were identified by the appearance of two bases at a given position in the sequence.
Phenotypes of Wild-Type and Mutant sad and dib Embryos.
Cuticle preparations, spectrin staining, and IMP-E1 expression in embryos have been described previously (6). See Fig. 5 and accompanying text, which are published as supporting information on the PNAS web site, www.pnas.org.
RIA.
Wild-type embryos (6–12 h old), or a mixed population of heterozygous and homozygous mutant sad embryos (6–12 h old) resulting from the mating of sad/TM3-arm-GFP reporter adults (6) or else selected homozygous sad embryos (10–14 h old) mechanically sorted for their lack of green fluorescent protein (GFP) fluorescence (Copas Select, Harvard Bioscience, Holliston, MA), were homogenized and extracted exhaustively with methanol. The pooled solvents were evaporated, and the residues, along with the selected residues of samples after their RP-HPLC or TLC purification, were subjected to RIA. The H22 antibody was used for embryo titers (13), and the SHO3 antibody for chromatography residues (14). Results are expressed in ecdysone equivalents.
Full-Length Coding Sequence for sad.
The entire cDNA sequence for sad was amplified from a Drosophila embryonic cDNA library in pNB40 (15) by PCR using a novel amplification protocol (16) that generates two complimentary strands of DNA containing the cDNA of interest and the plasmid. In this protocol, two pairs of primers were designed, primers A + B on the 5′ end and C + D on the 3′ end of the sad sequence: SadA, 5′-GATTCGCACCAGGAGCTGGTC-3′; SadB, 5′-CTTAGCCTGAGCCTCTTCCGTTC-3′; SadC, 5′-GCGACCAATGCAAGACCGCTG-3′; and SadD, 5′-CGTGTGGCACTCAAGCAGCTC-3′.
Whole-Mount RNA in Situ Hybridization of sad and dib.
Tissue fixation, RNA hybridization, and detection were done according to standard protocols (6). Sad sense and antisense riboprobes were synthesized from the NB40 plasmid containing the 1.8-kb sad cDNA full-length coding sequence and labeled with digoxigenin according to protocols for SP6 and T7 polymerases (Boehringer Mannheim).
An alternative protocol was used for obtaining the probe used for the in situ hybridization of second and third instar larval brain-ring gland complexes. Dib and sad PCR fragments were synthesized by using the following specific primers and late third instar larval total RNA: (dibUp1) GCGAAAAGACCAGAGTAACG; (dibDo1) CCCACAGCCTTTCAATCACA; (sadUp1) GTGCGAATCCTTAGCCTGAG; and (sadDo1) TACGCTGTCAACGGGCATCT. Each cDNA was purified, cloned into the PGEMT-easy vector (Promega), and labeled as above.
Transfection of S2 Cells.
S2 cells were transfected (17) with either the wild-type sad or dib (6) gene under the control of the actin 5C promoter or with a control construct constitutively expressing the GFP protein (18). Sad or dib cDNA was cloned as a HindIII/Not into the StuI/Not of the S2 expression vector pBRAcpA (19). The GFP expression construct was generated by subcloning GFP into pRactAdh.
Incubation of Transfected S2 Cells with Ecdysteroid Precursors.
Three days after the transfection of S2 cells with dib, sad, dib and sad together, or the GFP (control) expression vector constructs, the M3 medium from the cell (8 × 106) culture was replaced with fresh medium (1 ml) containing DMSO (1%), Tween 80 (0.001%), and the [3H]ketotriol or [3H]22dE (each 105 dpm, 1 nM) or the nonlabeled 2dE substrate (6.5 μg/ml, 14.5 μM) and incubated at 25°C for 2 h or 8 h. Various ecdysteroid standards (1 μg) were added to samples containing radiolabel before their exhaustive extraction with methanol. The pooled solvents were evaporated, the residues were taken up in 30% methanol, and the solutions were kept at −20°C until analyzed by RP-HPLC, TLC, and MS.
Chromatography.
Equipment and procedures for RP-HPLC and normal phase TLC have been described previously (8–12). For MS analysis, after sequential RP-HPLC and TLC purification of the sad 2dE metabolite, i.e., E, the residue was subjected to RP-HPLC (isocratic 15% acetonitrile)/electrospray MS (ESI-MS, Thermo Finnigan Quadrupole Electrospray, San Jose, CA).
Enzymatic Treatment of Ecdysteroid Metabolites.
After RP-HPLC and/or TLC analysis of tritiated metabolites from transfected S2 cell incubations, purified products were subjected to further biochemical modification by incubation with M. sexta day seven, fifth larval instar prothoracic gland homogenates (10), midgut microsomes (12), or Drosophila third larval instar brain ring gland complexes (9, 11) or adult ovaries (8). The extracted residues were analyzed by additional RP-HPLC and TLC.
Results
Confirmation of sad as CYP315A1.
Using information from the Drosophila genome project (http://flybase.bio.indiana.edu; ref. 20), we searched for a correlation between predicted chromosomal locations of CYP enzymes and the known genetic map positions for several Halloween group mutations. For the sad gene located at the 86F6–87A7 cytological interval of the third chromosome (7), we identified CYP315A1 (87A2) as a potential candidate CYP for the product of this locus. To confirm the tentative identity of sad, genomic DNA from two different sad mutant alleles were sequenced, and, in each case, a base alteration was identified that leads to formation of a stop codon. For sadZ309, glutamine 126 was converted to a stop codon whereas for sadZ804, tryptophan 442 was converted to a stop codon. Both mutations are presumed to be null alleles because each would eliminate the C-terminal positioned heme-binding site that is required for catalytic activity of all cytochrome-P450 enzymes.
Phenotype of Mutants.
See Fig. 5.
Embryonic Ecdysteroid Titers.
Ecdysteroid levels were decreased by 50% relative to wild type in the mixed 6- to 12-h embryo population (i.e., 25% homozygous sad, 50% heterozygous sad/balancer) collected from mated sad/TM3-arm-GFP adults, i.e., wild type = 54.28 ± 14.15 pg/mg wet weight (SD) vs. sad = 25.52 ± 9.89 pg/mg (P < 0.0004). Ecdysteroid titers in machine-selected homozygous sad embryos (10–14 h old) were below assay sensitivity using the H22 antibody, i.e., <1.6 pg/mg wet weight.
In Situ Expression of sad and dib.
To determine whether the sad gene product was a candidate for a modulator of E synthesis (Fig. 1) during development, the expression pattern of sad mRNA was analyzed during embryonic and postembryonic development and compared with that of dib. A full-length sad cDNA was used as a probe in Northern analysis, and the data revealed a single transcript of 1.8 kb at several developmental stages (data not shown). Using this cDNA as a riboprobe, in situ hybridization to blastoderm stage embryos demonstrated an early anterior enrichment of sad transcription (Fig. 2A) although much less than that found for dib (6). There was no sad mRNA in unfertilized eggs, suggesting that there is no maternal contribution of sad, as was noted for dib (6). By the time of complete germ band extension, sad expression was resolved into stripes, and, by stage 10–11, its expression was very reminiscent of a segment polarity gene, in that transcription occurred in a portion of the epidermis of every parasegment. Dib was also expressed in epidermal cells at this time, although its striped pattern is not as pronounced as that of sad (6). After stage 11, the expression of sad faded from all tissues and then resumed in the primordia of the ring gland at stage 15. This picture continued through the remainder of embryogenesis in the prothoracic gland cell component of the ring gland, and later within the larval ring gland. In adult females, both sad and dib transcripts are expressed strongly in the follicle cells, another site of ecdysteroid synthesis (2, 3, 5, 6). Unlike dib, however, sad is also expressed in the nurse cells.
Fig 1.
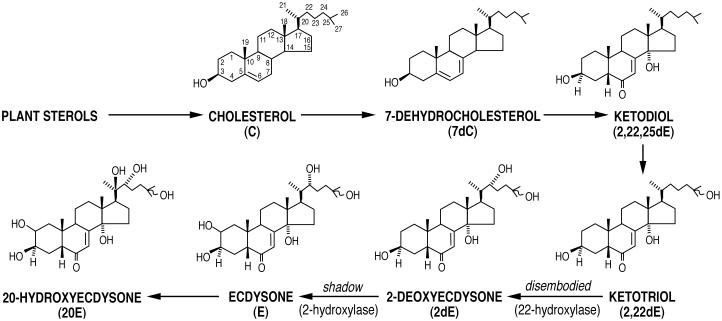
Theoretical scheme of ecdysone biosynthesis (2). Dietary cholesterol or phytosterols serve as the ultimate precursors for the synthesis of 20E, because insects cannot synthesize sterols from simple carbon molecules. The intermediate resulting from the subsequent oxidation of 7dC has not been identified unequivocally, but a number of in vivo and in vitro studies suggest that: (i) it is the ketodiol (2,22,25dE) or its 3-dehydro derivative; (ii) these complex reactions are catalyzed by one or more CYP enzymes present within the mitochondria; and (iii) the delivery of 7dC from the endoplasmic reticulum to these enzymes may be rate-limiting for the production of E. The sequential hydroxylations of the ketodiol at carbon 25 (to the ketotriol), carbon 22 (to 2dE by dib), carbon 2 (to E by sad), carbon 20 (activation to 20E), and carbon 26 (inactivation to 20,26E) are catalyzed by CYP enzymes present in the microsomal and/or mitochondrial compartments (2, 5).
Fig 2.
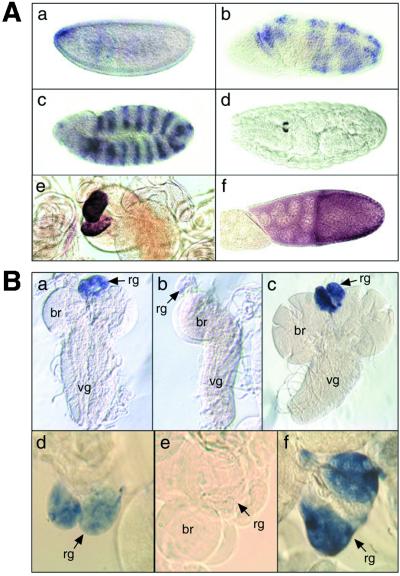
(A) In situ expression pattern of sad. (a) Cellular blastoderm stage embryo; (b) germ band extension stage 8, segmental staining appears in the epidermal cells; (c) stage 10–11 embryo showing segment polarity type expression; (d) ring gland expression in a stage 17 embryo; (e) expression in the prothoracic gland cells of the ring gland of a third instar larva; (f) stage 10 ovary. (B) Cycling of dib and sad expression between the second (L2) and third (L3) instar. In situ hybridization pattern of dib (a–c) and sad (d–f) within the prothoracic gland cells of the ring gland. (a and d) Late L2 larval brain-ring gland complex; (b and e) early L3 larval brain-ring gland complex; (c and f) late L3 larval brain-ring gland complex. Rg, Ring gland; br, brain; vg, ventral ganglion.
Because the ecdysteroid titer varies during the molting cycle (3), we determined whether the transcriptional activity of dib and sad correlates with these changes during larval development by examining the expression of dib and sad in the ring gland during the second and third larval instars (Fig. 2B). The data show that both dib and sad are expressed strongly in late second and third instar larvae. However, just after ecdysis to the third instar, dib and sad are down-regulated dramatically.
Characterization of the Roles of sad and dib.
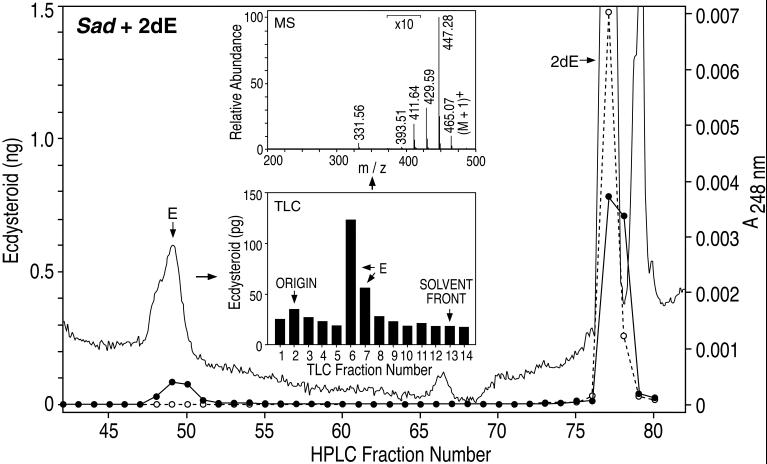
Three days after transfection with sad, S2 cells were incubated with 2dE (see Fig. 1) for 8 h, and the medium and cells were then extracted and analyzed by RP-HPLC/RIA (Fig. 3). The data reveal that S2 cells transfected with sad (CYP315A1) metabolize the 2dE substrate solely into E. No such activity was observed in GFP (control)-transfected cells. Over the 8-h incubation period, 0.25 μg E was recovered from the 6.5 μg 2dE added to the incubation mixture, a yield of 3.9% at a substrate concentration of 14.5 μM. Additional kinetic analysis revealed a Km of 10.3 μM and a Vmax of 1 nmol/8 h for this enzyme (data not shown). The characterization of this product as E was not only qualitatively consistent with its UV spectrum (UVmax 248 nm, data not shown), but also quantitatively consistent with both specific UV absorption and ecdysteroid immunoreactivity (Fig. 3). The identity of this product was confirmed by RP-HPLC/ESI-MS after additional normal phase TLC/RIA analysis (Fig. 3 Insets). Note the molecular ion (M + 1)+ at 465 and the sequential loss of four molecules of water at (M + 1)+-H2O(1–4), characteristic of E (21).
Fig 3.
RP-HPLC/TLC/RIA/MS analysis after sad- or GFP-transfected S2 cell incubation (8 h) with the 2dE substrate (30–100% methanol gradient). Ecdysteroid immunoreactivity was quantified by RIA (1/1000 of total sample) by using the SHO-3 antibody after incubations with sad (filled circles) or GFP (open circles). UV absorption was measured at 248 nm (solid line). (Inset, TLC) TLC/RIA (chloroform/ethanol) of RP-HPLC-purified ecdysone (E) product (1/1000 of total sample). (Inset, MS) RP-HPLC/ESI-MS on an LCQDECA (Thermo Finnigan, San Jose, CA) of the TLC-purified E product.
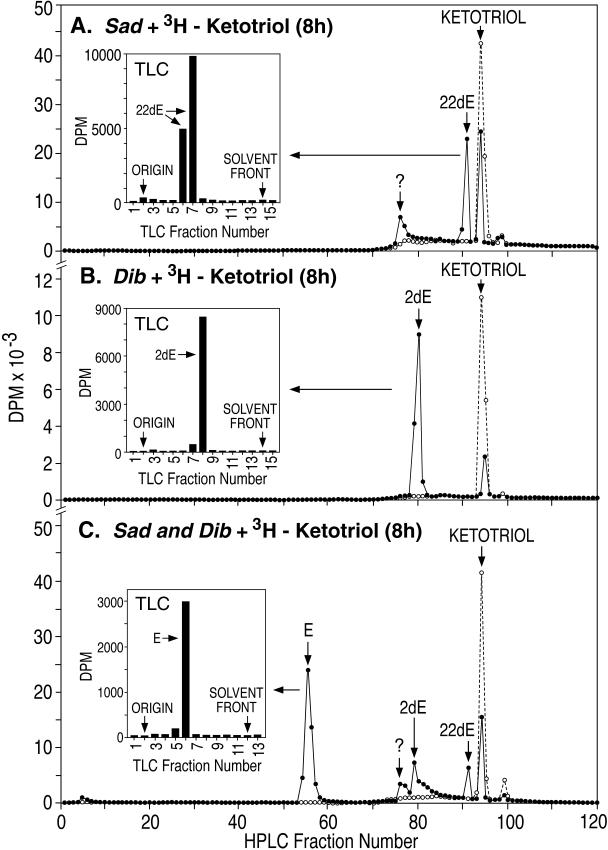
Further confirmation of ecdysteroid-specific 2-hydroxylase activity after sad expression in S2 cells was obtained on incubation of these cells with the [3H]ketotriol substrate, i.e., [3H]2,22-dideoxyecdysone ([3H]2,22dE; see Fig. 1). In this case, [3H]2,22dE (1 nM) was converted to [3H]22-dE (34.7%) and a conjugate of an unidentified metabolite (28.8%) over the 8-h incubation period, i.e., 63.5% total substrate metabolism (Fig. 4A; 14.4% after 2 h, data not shown). GFP (control)-transfected cells did not metabolize the [3H]ketotriol substrate significantly. This [3H]22dE product comigrated in TLC (Fig. 4A Inset) with the standard [3H]22dE metabolite produced from the [3H]ketotriol by enzymatic activity present in M. sexta prothoracic gland homogenates in vitro (12). We conclude from the data that sad (CYP315A1) codes for the ecdysteroid C2-hydroxylase (Fig. 1).
Fig 4.
RP-HPLC/TLC analysis of ecdysteroids after sad-, dib-, sad and dib-, or GFP-transfected S2 cell incubations with the [3H]ketotriol (2,22dE) substrate. (A) sad (filled circles) or GFP (open circles) + [3H]ketotriol. ?, Unidentified metabolite conjugate. (Inset) TLC analysis of [3H[22dE product. (B) dib (filled circles) or GFP (open circles) + [3H]ketotriol. (Inset) TLC of [3H]2dE product. (C) sad and dib (filled circles) or GFP (open circles) + [3H]ketotriol. (Inset) TLC of [3H]E (ecdysone) product.
To examine Dib function, we performed a similar RP-HPLC analysis (Fig. 4B). S2 cells transfected with dib were also able to metabolize this same [3H]ketotriol substrate, but, in this case, the sole product was [3H]2dE, i.e., 82.8% in 8 h (47.7% after 2 h, data not shown). The identity of this product was confirmed by its comigration in TLC with standard 2dE (Fig. 4B Inset) and its subsequent facile in vitro metabolism to E by M. sexta prothoracic glands (data not shown). As a result, we conclude that dib (CYP302A1) codes for the ecdysteroid C22-hydroxylase (Fig. 1).
As a final confirmation of these conclusions, S2 cells were cotransfected with both sad and dib and incubated with the [3H]ketotriol. In this case, the [3H]ketotriol substrate was metabolized efficiently to [3H]E in a 51.7% yield after 8 h of incubation (Fig. 4C). Product identity was confirmed when this [3H]E comigrated with standard E after TLC (Fig. 4C Inset). Additional proof of its identity was the demonstration of the facile conversion of this product to [3H]20E when incubated with a M. sexta larval midgut microsomal fraction exhibiting 20-monoxygenase activity (data not shown).
Additional experiments yielded information on the substrate specificity of Dib. Sad-transfected S2 cells could hydroxylate both 2dE and the [3H]ketotriol at carbon 2 (i.e., to E and [3H]22dE, respectively). However, when [3H]22dE was incubated subsequently with dib-transfected cells, no further metabolism was observed. This intermediate was also not metabolized further in vitro by larval brain-ring gland complexes or adult ovaries (data not shown). Apparently, 22dE is not a substrate for the 22-hydroxylase coded for by dib, either in transfected S2 cells or within Drosophila endocrine organs expressing dib and sad.
Discussion
In contrast to the identification of dib by classical molecular techniques (6), a preliminary identification of sad was achieved by comparing its known genetic map position (86F6–87A7) to predicted chromosomal locations of CYP enzymes by using information from the Drosophila genome project, i.e., CYP315A1 at 87A2. This initial correlation was confirmed by genomic PCR and sequencing when it was found that base alterations leading to premature stop codons created null alleles lacking the critical catalytic heme-binding site. The insect genes most closely related to sad and dib are CYP12A1, a mitochondrial P450 from the house fly (22), and CYP12B1 from Drosophila acanthoptera (23). As with these enzymes, Sad contains a conserved N-terminal amphipathic region required for mitochondrial import (24). Indeed, colocalization of a mitochondrial marker with epitope-tagged versions of Dib and Sad was observed after transient transfection in S2 cells (G.M., A.P., and M.B.O., unpublished data).
The characterization of Sad and Dib as the first CYP enzymes shown definitively to be directly involved in ecdysone biosynthesis was achieved after their transfection into Drosophila S2 cell cultures derived from embryos. Incubation of these cells with radiolabeled and nonlabeled ecdysteroid precursors demonstrated conclusively that Dib and Sad catalyze the terminal hydroxylations of the ecdysteroid molecule at the C22 and C2 positions, respectively (Fig. 1). This identity was confirmed when S2 cells transfected with both dib and sad were able to metabolize the ecdysteroid precursor [3H]2,22dE to [3H]E, in exactly the same manner as occurs when this substrate is incubated with Drosophila ring glands in vitro (8, 9, 11) or administered in vivo (J.T.W., unpublished data).
The mutant embryonic phenotype of sad closely resembles that of dib (ref. 6 and see Fig. 5), most likely a result of the very low ecdysteroid titers in these embryos. In situ expression of the two genes during Drosophila development is also quite similar (Fig. 2). Understanding both the temporal and tissue-specific regulation of the synthesis of these P450 enzymes should elucidate the control of ecdysone biosynthesis as well as provide new insights into Drosophila embryonic and postembryonic development per se. For example, ecdysteroid levels begin to rise during early embryogenesis at about the time of gastrulation (stage 6–7, 3 h after egg laying) and peak at stage 11–12 (7–9 h after egg laying) during germ band shortening (25), i.e., about the time when sad expression is strongly localized in the region of epidermal stripes. This result occurs hours before the formation of the embryonic ring gland at stage 15 (11–13 h after egg laying), although the first appearance of progenitor prothoracic gland cells derived from the dorsal wall of the invaginating stomodaeum has been noted at 9–10 h (26). The striped pattern of sad expression is particularly notable in light of recent findings that retinoid-elicited patterning along the vertebrate anterior-posterior axis depends on the different spatial distributions of the P450-type enzymes involved in retinoid biosynthesis and degradation (27–30). By such source and sink mechanisms, gradients of hormone may be established in regional specific ways (31), which in turn could influence tissue development.
These findings suggest strongly that the expression of sad, like dib, is absolutely required for the embryonic production of E. In fact, the data support the hypothesis that ecdysteroids are synthesized de novo during Drosophila embryogenesis, perhaps within the epidermal segments. That is, a sterol or ecdysterol precursor which at the least lacks the 22- and 2-hydroxyl functions of E (see Fig. 1), is converted to ecdysteroid by the sequential action of Dib and Sad. The role of maternal RIA-positive ecdysteroid conjugates (32–34) and their possible contribution to this mid-embryonic surge awaits future research. Nevertheless, during larval development, the level of sad and dib transcripts within the prothoracic gland cells of the ring gland appear to cycle in concert with increases in the premolt hemolymph ecdysteroid titer (35, 36) at the end of both the second and third instars (Fig. 2B). In addition, both dib and sad are completely down-regulated between the instars. These initial observations provide the base necessary for critical analysis of the control of hormone levels in a genetic system.
The present data indicate that two key mitochondrial CYP enzymes that participate in E biosynthesis have been identified and suggest that our approach will facilitate the characterization of the remaining unidentified P450 enzymes implicated in the ecdysteroidogenic pathway (Fig. 1). Indeed, we have found recently that the Halloween genes phantom, shade, and spook also exhibit defects in ecdysteroid biosynthesis and all code for CYP300 level enzymes, but of the microsomal class (A.P., J.T.W., L.I.G., and M.B.O., unpublished data). As such, they likely function before the action of dib and sad. The complete characterization of their gene products and those of other P450 genes in the pathway leading to 20E should set the stage for understanding the biochemical and molecular control of steroid molting hormone production in this dipteran model. Finally, because all insects require ecdysteroids for proper development, these specific CYP enzymes may provide novel targets for the development of agents that can be used for the control of insects of both agricultural and biomedical importance.
Supplementary Material
Acknowledgments
We thank Sue Whitfield for graphics and Hasan Koc for mass spectrometric analysis. The SHO-3 antibody was a gift from Dr. Sho Sakurai (Kanazawa University, Japan). The Drosophila embryonic cDNA library was a gift of Nick Brown (Cambridge University, U.K.). Plasmids pRactAdh and pBRAcpA were provided by Dr. Lucy Cherbas (University of Indiana). The University of North Carolina MS facility is supported in part by National Institutes of Health (NIH) Grants P30 CA16086 and P30 ES10126. This work was supported by National Science Foundation Grant IBN0130825 (to L.I.G. and J.T.W.). A.P. was supported by NIH Training Grant HD33692. M.B.O. is an Associate Investigator of the Howard Hughes Medical Institute.
Abbreviations
2dE, 2-deoxyecdysone
22dE, 22-deoxyecdysone
2,22dE, 2,22-dideoxyecdysone (ketotriol)
dib, disembodied
E, ecdysone
20E, 20-hydroxyecdysone
sad, shadow
CYP, cytochrome P450 monooxygenase
GFP, green fluorescent protein
ESI, electrospray ionization
Data deposition: The sequence reported in this paper has been deposited in the GenBank database [accession no. AY079170 (sad)].
References
- 1.Manglesdorf D. J., Thummel, C., Beato, M., Herrlich, P., Schutz, G., Umesono, K., Blumberg, B., Kastner, P., Mark, M. & Chambon, P. (1995) Cell 83, 835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilbert L. I., Rybczynski, R. & Warren, J. T. (2002) Annu. Rev. Entomol. 47, 883-916. [DOI] [PubMed] [Google Scholar]
- 3.Henrich V. C., Rybczynski, R. & Gilbert, L. I. (1999) in Vitamins and Hormones, ed. Litwack, G. (Academic, San Diego), Vol. 55, pp. 73–125. [DOI] [PubMed] [Google Scholar]
- 4.Thummel C. S. (1995) Cell 83, 871-877. [DOI] [PubMed] [Google Scholar]
- 5.Kappler C., Kabbouh, M., Hetru, C., Durst, F. & Hoffmann, J. (1988) J. Steroid Biochem. 31, 891-898. [DOI] [PubMed] [Google Scholar]
- 6.Chávez V. M., Marquès, G., Delbecque, J. P., Kobayashi, K., Hollingsworth, M., Burr, J., Natzle, J. & O'Connor, M. (2000) Development (Cambridge, U.K.) 127, 4115-4126. [DOI] [PubMed] [Google Scholar]
- 7.Jürgens G., Wieschaus, E., Nüsslein-Volhard, C. & Kluding, C. H. (1984) Roux's Arch. Dev. Biol. 193, 283-295. [DOI] [PubMed] [Google Scholar]
- 8.Warren J. T., Bachman, J., Dai, J.-D. & Gilbert, L. I. (1996) Insect Biochem. Mol. Biol. 26, 931-943. [DOI] [PubMed] [Google Scholar]
- 9.Warren J. T., Wismar, J., Subrahmanyam, B. & Gilbert, L. I. (2001) Mol. Cell. Endocrinol. 181, 1-14. [DOI] [PubMed] [Google Scholar]
- 10.Warren J. T. & Gilbert, L. I. (1996) Insect Biochem. Mol. Biol. 26, 917-929. [DOI] [PubMed] [Google Scholar]
- 11.Warren J. T., Dai, J.-D. & Gilbert, L. I. (2000) Insect Biochem. Mol. Biol. 29, 571-579. [DOI] [PubMed] [Google Scholar]
- 12.Grieneisen M. L., Warren, J. T. & Gilbert, L. I. (1993) Insect Biochem. Mol. Biol. 23, 13-24. [DOI] [PubMed] [Google Scholar]
- 13.Warren J. T. & Gilbert, L. I. (1988) in Immunological Techniques in Insect Biology, eds. Gilbert, L. I. & Miller, T. M. (Springer, Heidelberg), pp. 181–214.
- 14.Kiriishi S., Rountree, D., Sakurai, S. & Gilbert, L. I. (1990) Experientia 46, 716-721. [DOI] [PubMed] [Google Scholar]
- 15.Brown N. H. & Kafatos, F. C. (1988) J. Mol. Biol. 203, 425-437. [DOI] [PubMed] [Google Scholar]
- 16.Haerry T. & O'Connor, M. (2002) Gene 291, 85-93. [DOI] [PubMed] [Google Scholar]
- 17.Ross J. J., Shimmi, O., Vilmos, P., Petryk, A., Kim, H., Gaudenz, K., Hermanson, S., Ekker, S., O'Connor, M. & Marsh, J. (2001) Nature (London) 410, 479-483. [DOI] [PubMed] [Google Scholar]
- 18.Bhanot P., Brink, M., Samos, C., Hsieh, J., Wang, Y., Macke, J., Andrew, D., Nathans, J. & Nusse, R. (1996) Nature (London) 382, 225-230. [DOI] [PubMed] [Google Scholar]
- 19.Cherbas L., Moss, R. & Cherbas, P. (1994) Methods Cell. Biol. 44, 161-179. [DOI] [PubMed] [Google Scholar]
- 20.Adams M. D., Celniker, S., Holt, R., Evans, C., Gocayne, J., Amanatides, P., Scherer, S., Li, P., Hoskins, R., Galle, R., et al. (2000) Science 287, 2185-2195. [DOI] [PubMed] [Google Scholar]
- 21.Wainwright G., Prescott, M., Lomas, L., Webster, S. & Rees, H. H. (1997) Arch. Insect Biochem. Physiol. 35, 21-31. [Google Scholar]
- 22.Guzov V., Unnithan, G., Chernogolov, A. & Feyereisen, R. (1998) Arch. Biochem. Biophys. 359, 231-240. [DOI] [PubMed] [Google Scholar]
- 23.Danielson P. B. & Fogleman, J. C. (1997) Insect Biochem. Mol. Biol. 27, 595-604. [DOI] [PubMed] [Google Scholar]
- 24.Tiijet N., Helvig, C. & Feyereisen, R. (2001) Gene 262, 189-198. [DOI] [PubMed] [Google Scholar]
- 25.Maroy P., Kaufmann, G. & Dübendorfer, A. (1988) J. Insect Physiol. 34, 633-637. [Google Scholar]
- 26.Poulson D. F. (1945) Trans. Conn. Acad. Arts Sci. 36, 449-487. [Google Scholar]
- 27.de Roos K., Sonneveld, E., Compaan, P., ten Berge, D., Durston, A. & van der Saag, P. (1999) Mech. Dev. 82, 205-211. [DOI] [PubMed] [Google Scholar]
- 28.Swindell E. C., Thaller, C., Sockanathan, S., Petkovich, M., Jessell, T. & Eichele, G. (1999) Dev. Biol. 216, 282-296. [DOI] [PubMed] [Google Scholar]
- 29.Hollemann T., Chen, Y., Grunz, H. & Pieler, T. (1998) EMBO J. 17, 7361-7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCaffery P., Wagnerl, E., O'Neill, J., Petkovich, M. & Drager, U. C. (1999) Mech. Dev. 85, 201-203. [DOI] [PubMed] [Google Scholar]
- 31.Stoilov I. (2001) Trends Genet. 17, 629-632. [DOI] [PubMed] [Google Scholar]
- 32.Bownes M., Shirras, A., Blair, M., Collins, J. & Coulson, A. (1988) Proc. Natl. Acad. Sci. USA 85, 1554-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grau V. & Gutzeit, H. O. (1990) Roux's Arch. Dev. Biol. 198, 295-302. [DOI] [PubMed] [Google Scholar]
- 34.Grau V., Píš, J. & Lafont, R. (1995) Eur. J. Entomol. 92, 189-196. [Google Scholar]
- 35.Maroy P., Koczka, K., Fekete, E. & Vargha, J. (1980) Drosophila Inf. Serv. 55, 98-99. [Google Scholar]
- 36.Richards G. (1981) Mol. Cell. Endocrinol. 21, 181-197. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.