Abstract
The structure of the reaction center from Rhodobacter sphaeroides has been solved by using x-ray diffraction at a 2.55-Å resolution limit. Three lipid molecules that lie on the surface of the protein are resolved in the electron density maps. In addition to a cardiolipin that has previously been reported [McAuley, K. E., Fyfe, P. K., Ridge, J. P., Isaacs, N. W., Cogdell, R. J. & Jones, M. R. (1999) Proc. Natl. Acad. Sci. USA 96, 14706–14711], two other major lipids of the cell membrane are found, a phosphatidylcholine and a glucosylgalactosyl diacylglycerol. The presence of these three lipids has been confirmed by laser mass spectroscopy. The lipids are located in the hydrophobic region of the protein surface and interact predominately with hydrophobic amino acids, in particular aromatic residues. Although the cardiolipin is over 15 Å from the cofactors, the other two lipids are in close contact with the cofactors and may contribute to the difference in energetics for the two branches of cofactors that is primarily responsible for the asymmetry of electron transfer. The glycolipid is 3.5 Å from the active bacteriochlorophyll monomer and shields this cofactor from the solvent in contrast to a much greater exposed surface evident for the inactive bacteriochlorophyll monomer. The phosphate atom of phosphatidylcholine is 6.5 Å from the inactive bacteriopheophytin, and the associated electrostatic interactions may contribute to electron transfer rates involving this cofactor. Overall, the lipids span a distance of ≈30 Å, which is consistent with a bilayer-like arrangement suggesting the presence of an “inner shell” of lipids around membrane proteins that is critical for membrane function.
Integral membrane proteins are found in the cell membrane surrounded by lipids. The biophysical and biochemical properties of these proteins are critically determined by the lipid environment. The lipid composition of cell membranes is complex, containing a variety of phospholipids, glycolipids, and other small molecules. In the purple nonsulfur bacterium Rhodobacter sphaeroides, the major lipids are the phospholipids, phosphatidylcholine, phosphatidylethanolamine, phosphatidylglycerol, and cardiolipin, or diphosphatidyl glycerol, and two glycolipids, sulfoquinovosyl diacylglycerol and glucosylgalactosyl diacylglycerol (1). The relative amounts of these lipids vary significantly depending on specific growth conditions, in particular the oxygen level, although the fatty acid chains are always predominately 18:1 with only minor contributions from 18:0, 16:0, and 16:1 and trace amounts of smaller chains. The lipid composition is known to critically determine the morphology of the cell membrane, but whether the lipids can affect the photosynthetic energy conversion processes remains an outstanding question.
Understanding the influence of the lipid properties on the photosynthetic complexes, or any other integral membrane proteins, is in most cases limited by the lack of detailed structural information concerning membrane proteins. To solve the three-dimensional structure of an integral membrane protein, the protein must first be solubilized with detergent or lipids at protein concentrations that are sufficient to reach supersaturation and drive the nucleation process. Because of the additional parameters introduced by the detergent and amphiphile molecules, the crystallization process of membrane proteins is complex (2), and the number of structures of membrane proteins that have been determined is limited.
Among the best determined structures of an integral membrane protein are the bacterial reaction centers (RCs; refs. 3 and 4). In many of these models, detergent molecules were positioned to accommodate extended regions of electron density found on the surface of the protein. As the quality of the x-ray diffraction data improved, one of these detergent molecules was identified as a bound cardiolipin (5). This cardiolipin is quite removed from the cofactors and has no identifiable effect on the electron transfer properties of the protein. In this manuscript we report the resolution of additional areas of electron density that are assigned to two lipid molecules on the surface of the RC from R. sphaeroides. The identity of these lipids is confirmed by using laser mass spectroscopy. The implications of these lipids on the function of the cofactors, the protein interactions that lead to lipid binding, and the properties of the protein in the membrane are discussed.
Materials and Methods
Purification and Crystallization of RCs.
The wild-type RCs were isolated from a deletion strain complemented with the wild-type genes (6). Cells were grown semiaerobically under nonphotosynthetic conditions, and RCs were isolated after published conditions by using lauryl dimethylamine oxide (Fluka) as previously described (7). Crystals were grown with sizes up to 10 mm in length by using conditions similar to those described by Ermler and coworkers (8). Briefly, a sitting drop setup was used at room temperature with a protein solution consisting of 3.5% heptano-1,2,3-triol, 0.1% lauryl dimethylamine oxide, 2.0% dioxane, and 0.75 M potassium phosphate (pH 7.4). Droplets of 50 μl with a final protein concentration of 0.12 mM were equilibrated against a 1-ml reservoir solution made with 1.6 M potassium phosphate buffer (pH 7.4).
Data Collection.
Crystals were initially measured at room temperature on a Rigaku (Tokyo) R-AXIS IIC image plate detector mounted on a Rigaku RU-200HB copper rotating anode x-ray generator. The data were processed and scaled by using denzo and scalepack (9). No serious radiation damage was detected during the data collection in any of the crystals. Data to the higher resolution limit of 2.55 Å were subsequently collected by using synchrotron radiation with a wavelength of 1.08 Å at beamline 7-1 of the Stanford Synchrotron Radiation Laboratory using a MAR Research Image Plate. The synchrotron data were processed by using the mosflm package (10) and scaled with scala (11).
Refinement and Model Building.
Crystallographic refinement was performed by using the cns package (12) and modeled by using o (13). Refinement was initiated by using RC coordinates of the trigonal form (5) (PDB accession number ) with the appropriate amino acid substitution at the mutated site of that mutant structure. Rigid body refinement was performed before positional refinement by using 3-Å resolution data. Cycles of positional refinement and temperature factor refinement were alternated with manual building by using σA-weighted (2|FO| − |Fc|) and (|FO| − |Fc|) electron density maps. The last cycles of refinement were performed with 2.55-Å resolution data. Possible water molecules were identified by using waterpick in cns (12) and retained in the model provided they were located in positions of positive peaks in (|FO| − |Fc|) maps and in a suitable hydrogen-bonding environment. The resulting model was then refined, and water molecules with B factors higher than 70 Å2 were discarded. The quality of the final model was analyzed by using the program procheck (14). Some of the figures were made by using the program xtalview (15).
Lipid Analysis by Matrix-Assisted Laser Desorption Ionization (MALDI).
The identities of the lipids bound to the protein were determined by measuring their molecular masses using MALDI time-of-flight mass spectrometry (Vestec Lasertec Research, Houston). A chloroform-methanol (1:1) extract of the RC preparation was mixed with the matrices 4-hydroxybenzylidenemalononitritrile‡ and terthiophene (16) in the same solvent. One-microliter aliquots were dried on stainless steel sample pins for measurements performed in both positive and negative ion modes. The matrix solution was mixed with an equal volume of 50 micromolar RC, and 1-μl aliquots were dried on stainless steel pins for measurement. Calibration was performed only in the positive ion mode by using two synthetic polypeptides with masses of 568.7 and 1,980.2 Da desorbed from α-cyano-4-hydroxycinnamic acid as external standards. Because of the lack of an internal standard and the use of a different matrix during calibration, the absolute values of the masses may be inaccurate by up to 2 Da although the relative values are accurate to a few tenths of a dalton.
Results
X-Ray Diffraction Data.
Crystals were obtained in the trigonal space group P3121 with cell constants a = b = 141.8 Å and c = 187.5 Å as previously reported for wild type (8). The initial x-ray data used for the analysis were collected in the laboratory and extended to a 3.4-Å resolution limit. The quality of the data was greatly improved by measuring the diffraction at the Stanford Synchrotron Radiation Laboratory. These experiments yielded a total of 564,825 observations and 69,691 unique reflections, which were integrated with an Rmerge of 12.6% representing 97% of all possible reflections (Table 1).
Table 1.
Summary of diffraction and structural data
| Space group | P3121 |
| Cell parameters a, b, c, Å | 141.8, 141.8, 187.5 |
| Resolution range, Å | 30.0–2.55 |
| Total reflections | 564, 825 |
| Unique reflections | 69, 691 |
| I/σ | 4.3 (1.5) |
| Rmerge, % | 12.6 (40.6) |
| Completeness, % | 97.2 (92.5) |
| Rcryst, Rfree, % | 18.3, 20.5 |
| rms bond deviations, Å | 0.010 |
| rms angle deviations, ° | 1.63 |
| Ramachandran plot | |
| Most favorable, % | 91.4 |
| Additionally allowed, % | 8.6 |
| Not allowed, % | 0.0 |
| Average B-factor, Å2 | 36.5 |
Overall data with last 0.1 Å shell in parentheses.
Structural Model of the Protein Subunits and Cofactors.
The model is in excellent agreement with the diffraction data as the final R factor is 18.3% and the Rfree is 20.5%. The geometry of the resulting structure is also excellent and the Ramachandran plot has almost all of the residues located in the most favorable region, with the remaining few found in the additional allowed regions. None of the residues are located in the disallowed region.
The model is composed of three protein subunits consisting of all 281 residues for the L subunit, 302 residues for the M subunit with the carboxyl terminal residues 303–307 omitted, and 239 residues for the H subunit, with residues 1–10 and the carboxyl terminal residues 249–260 omitted. The cofactors all fit the electron density very well except for the secondary quinone, QB. As reported for other models based on the trigonal crystal form, low density was found for QB. To fit the occupancy of this ubiquinone, all of the B-factors were set to 40 Å2, the approximate average B-factor for the structure, and QGROUP refinement was made by using the program cns. The resulting QB occupancy was estimated to be less than 40%. Possible reasons for this low occupancy have been addressed by other authors (17). Moreover, as demonstrated for a model based on the tetragonal form, QB can be positioned in two alternative conformations corresponding to the RCs in dark and light conformations (18). These alternative positions are also evident in models derived from the trigonal crystal form (5). In our (2|FO| − |Fc|) electron density maps, the density at the QB pocket is too weak to distinguish between the two possible locations for the QB ring. Also, the isoprenoid chain could not be unambiguously traced and is truncated in the final model.
Structural Model of the Bound Lipids.
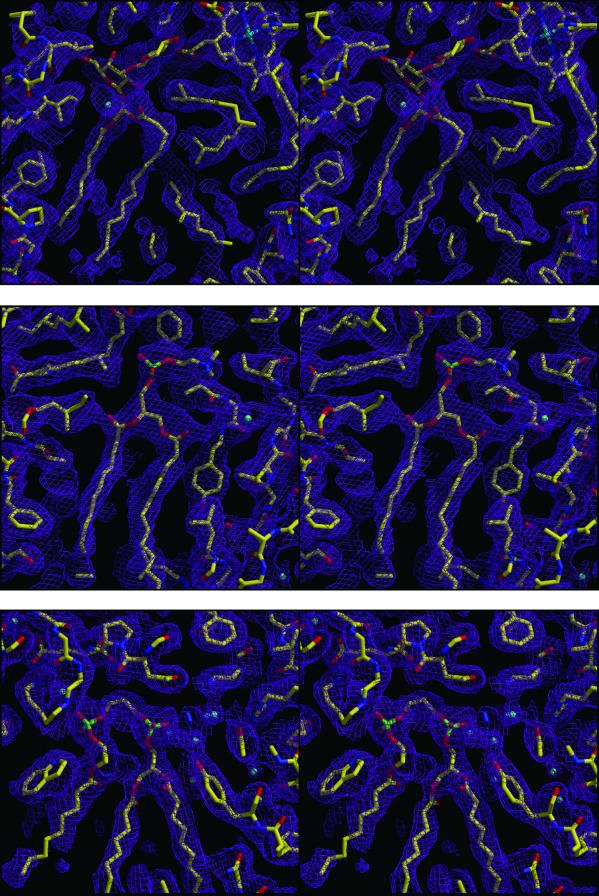
The electron density map shows the presence of three different lipids on the surface of the protein (Fig. 1). Near the active branch of the cofactors is an extended feature of electron density that can be well fitted by using the glycolipid glucosylgalactosyl diacylglycerol (Fig. 1 Top). Toward the cytoplasmic side of the protein on the surface of the M subunit at a distance of approximately 10 Å from the ring of QB, there is an extended electron density that can be well fitted with phosphatidylcholine (Fig. 1 Middle). The polar group of the lipid is near the inactive bacteriopheophytin with the phosphate atom being ≈6.5 Å from the conjugated ring. A third lipid, a cardiolipin, is located on the M subunit at the position previously reported (Fig. 1 Bottom; ref. 5). The electron density for this lipid is notably extensive because of the presence of the four fatty acid chains in contrast to the two chains present for the other two lipids.
Fig 1.
Stereoview of electron density maps (2|FO| − |Fc|) for the three bound lipids (Top) glucosylgalactosyl diacylglycerol, (Middle) phosphatidylcholine, (Bottom) cardiolipin and the surrounding protein residues. The identification of the glycolipid and cardiolipin is clear based on the electron density. However, the phosphatidylcholine can also be substituted with the slightly smaller phosphatidylethanolamine. The contour level is 1 σ.
For all three lipids, inclusion of the lipid slightly improves the R factor. The electron density for the polar group of each lipid is the best resolved region with the electron density being weaker for the hydrocarbon chains, particularly those that have limited interactions with the protein. Based on the electron density, the lipids have oleyl chains rather than a shorter length chain. The type of lipid can be assigned from the electron density although the precise chemical nature of the polar groups cannot be uniquely defined. For example, the density near QB can be modeled as either a phosphatidylcholine or a phosphatidylethanolamine. The electron density for the glycolipid is well defined for the polar head, identifying it as a lipid possessing a disaccharide moiety. The cardiolipin is distinctive because of the presence of the four alkyl chains.
Lipid Identification by MALDI.
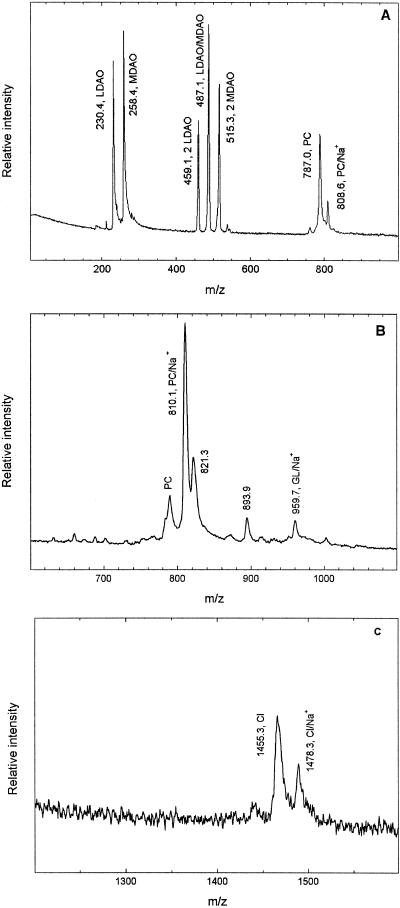
To determine which specific lipids are present, laser mass spectroscopy was performed on preparations of RCs that were also used to grow crystals. Because the lipids have different chemical natures, the measurements were performed by using different matrices in both negative and positive ion modes. These data strongly support the presence of three lipids bound to the RCs. The most pronounced peaks arise from the phospholipid dioleyl phosphatidylcholine, which has a molecular mass of 786.1 Da and is present only in the positive ion mode as expected (Fig. 2A). There are some weak peaks in some of the spectra that are probably due to dioleyl phosphatidylethanolamine, which has a molecular mass of 744.0 Da (data not shown). The peak amplitudes for this lipid are smaller than those for phosphatidylcholine, and these two lipids are probably both present at this site, with the phosphatidylcholine being significantly more abundant. The glycolipid lipid is not charged, and so the amplitudes of its peaks are intrinsically weak except for the sodium-adduct form, which has a net molecular mass of 957.1 Da (Fig. 2B). To verify the identify of this Na+ adduct, the concentration of sodium iodide in the matrix was varied from 0 to 25 mM, and the amplitude of this peak was found to systematically increase as the amount of sodium iodide increased. Control experiments with the commercially available glycolipid digalactosyl diglygeride (Sigma) revealed a similar effect of sodium iodide addition (data not shown). The peak corresponding to tetraoleyl cardiolipin is somewhat weak compared with the others because the larger size of 1,455.9 Da makes the cardiolipin more difficult to get into the gas phase than the smaller lipids (Fig. 2C).
Fig 2.
Representative spectra of purified RCs from laser mass spectroscopy using different matrices and ion modes. (A) Peaks at 787.0 and 808.6 Da correspond to the calculated molecular masses of phosphatidylcholine (PC), 786.1 Da, the sodium adduct (PC/Na+), 808.6 Da. The lower mass peaks at 230.4 and 258.4 Da are due to the detergent and show a mixture of lauryl and myrstyl dimethylamine oxide (LDAO and MDAO) that have calculated molecular masses of 230.4 and 258.4 Da, respectively. Also evident are dimers of lauryl and myrstyl dimethylamine oxide at 459.1 and 515.3 Da, as well as a lauryl/myrstyl dimethylamine oxide heterodimer at 487.1 Da. This spectrum was taken by using 4-hydroxybenzylidenemalononitritrile in positive ion mode. (B) A peak at 959.7 Da corresponding to the sodium adduct of the glycolipid (GL/Na+) with a calculated mass of 957.1 Da is evident using this 4-hydroxybenzylidenemalononitritrile matrix. The amplitude of this peak was found to track with the amount of NaI added to the matrix. Also seen in the spectrum is a major contribution from a sodium adduct of phosphatidylcholine at 810.1 Da. A minor contribution from the monoprotonated form is seen near 786 Da although the peak position is not precisely determined because of the presence of an overlapping peak. The peak at 893.9 Da arises from a contaminant. (C) Peaks at 1,455.3 and 1,478.3 Da corresponding to monoprotonated and monosodiated cardiolipin anions (CL and CL/Na+) with calculated molecular masses of 1,455.9 and 1,477.9 Da, respectively. This spectrum was taken by using a terthiophene matrix in negative ion mode.
Other peaks found in the spectra were easily assigned to known chemical species, such as the cofactors and detergent. For example, the cofactors are evident in spectra of samples measured by using a terthiophene matrix in the positive ion mode (data not shown). Interestingly, the spectra (Fig. 2A) indicate that the detergent is composed of approximately equal amounts of lauryl and myrstyl dimethylamine oxide although the solutions were made by using a pure commercial grade of lauryl dimethyl amineoxide. The peaks resulting from the two detergents were also present in spectra of solutions containing only the buffer and detergent (data not shown). The effect of this mixture of detergents on the crystallization is under investigation. Based on the amplitudes of the remaining peaks, other lipids may also be present but in much smaller amounts than those identified.
Discussion
The three-dimensional structure has been determined for the RC from R. sphaeroides. The accuracy of this model minimizes error in the electron density maps and provides the opportunity to model features of the protein surface that are partially disordered and consequently weak in the electron density maps. Three lipid molecules, namely a phosphatidylcholine, a cardiolipin, and a glycolipid, have been identified in the electron density, and the identity of these lipid molecules was confirmed by laser mass spectroscopy. The lipid binding sites are discussed, and this structural model is compared with previous models of the RC with an emphasis on the surface features of those models. The implications of these bound lipids on the electron transfer properties are discussed. Finally, a “quasiprotein” model is proposed for the properties of integral membrane proteins in cell membranes.
Lipid Binding Sites.
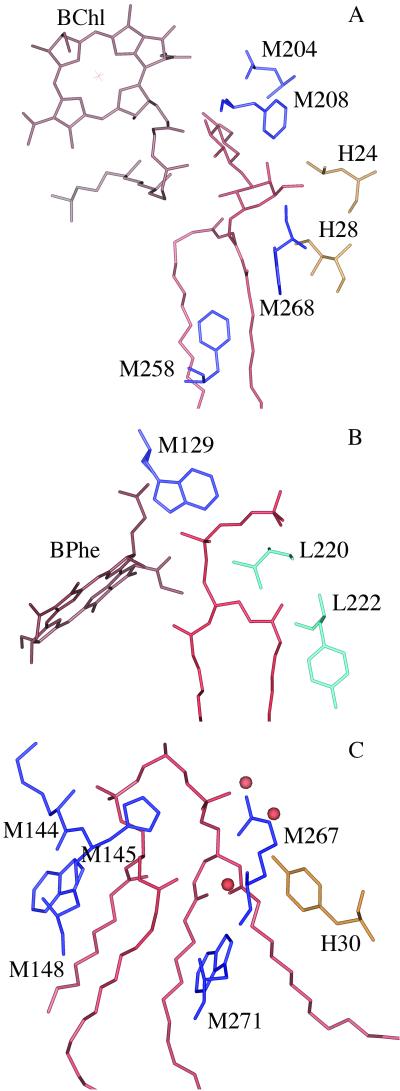
Three lipid molecules were found to bind to the surface of the protein at three distinct sites. The glycolipid interacts primarily with the M subunit and the transmembrane helix of the H subunit with the disaccharide moieties near the active bacteriochlorophyll monomer (Fig. 3A). The rings of the lipid come within 4.5 Å of the esters of ring E and 5.5 Å to the conjugated macrocycle. The lipid chains make contact with the isoprenoid chain of QA and Phe M258; this cofactor is lost and the region of the protein interacting with the fatty acid chain is significantly altered in the mutant structure of McAuley and coworkers (5). The phosphatidylcholine binds at the interface between the L and M subunits with the phosphate near the inactive bacteriopheophytin (Fig. 3B). The phosphate atom of the lipid lies 6.5 Å from the conjugated ring of the macrocycle. The ring of the secondary quinone lies 9.5 Å from the lipid but the isoprenoid chain comes in contact with the lipid. The polar group of the cardiolipin is over 15 Å from any of the cofactors and is stabilized by interactions with Arg M267 and His M145, as well as several water molecules as reported earlier (Fig. 3C). For all three lipids, the fatty acid chains make contacts with predominantly hydrophobic residues, especially aromatic residues and leucines. Thus, whereas the cardiolipin is largely exposed and binds tightly because of ionic interactions, the interactions involving the glycolipid and phosphatidylcholine involve primarily hydrophobic interactions as they are more buried in the protein (Fig. 4).
Fig 3.
Structure of the lipids with nearby cofactors and the surrounding protein. (A) The glycolipid (red) binds near the active bacteriochlorophyll monomer (brown) and nearby amino acid residues of the M subunit (blue) Leu M204, Phe M208, Phe M258, and Trp M268, and the H subunit residues (yellow) Leu H24 and Ile H28. (B) The phospholipid phosphatidylcholine (red) binds near the L subunit residues (light blue) Val L220 and Tyr L222, and the M subunit residue Trp M129 (dark blue), and the inactive bacteriopheophytin (brown). (C) The cardiolipin (red) binds on the M subunit (dark blue) and near the H subunit (yellow) with the phosphate atoms of the lipid interacting with His M145, Arg M267, Tyr H30, and several water molecules (orange spheres). Also shown are Lys M144, Trp M148, and Trp M271.
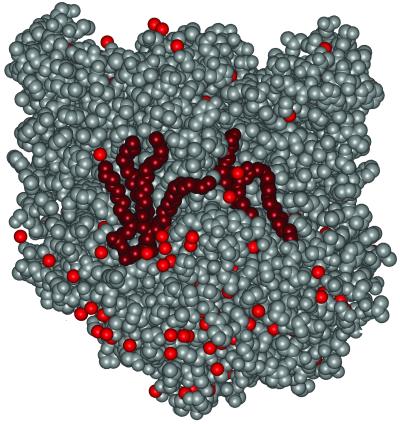
Fig 4.
Surface representation of the RC structure showing the glycolipid and cardiolipid (dark red), the protein (gray), and bound water molecules (light red). Both of the lipids are tightly bound, with the glycolipid fitting between the M and H subunits whereas the cardiolipin is more exposed but held by electrostatic interactions involving the two phosphates and nearby charged residues. The two lipids are positioned with a separation of about 30 Å between the phosphate and saccharide moieties.
Comparison with Earlier RC Models.
The overall structure of the protein is very similar to previous models of the RC from R. sphaeroides (5, 8, 18–20), with no significant alterations of the cofactor organization or interactions between the cofactors and protein side chains. However, unlike previous models, the current model does include three bound lipids on the surface of the protein. In earlier models of the protein, these regions have been identified as arising from bound detergent molecules (8, 18). For example, the glycolipid was previously modeled as arising from bound detergent, specifically three lauryl dimethylamine oxide molecules. The use of the higher quality electron density maps shows that the density in this region is continuous and suitable for the lipid. The electron density for the phosphatidylcholine also has been modeled as arising from a lauryl dimethylamine oxide molecule, although interpretation of this region is strongly influenced also by the positioning of the phytyl and isoprenoid chains of the cofactors. The electron density for the cardiolipin region was previously modeled as arising from a phosphate ion at the position of one of the phosphate moieties and a lauryl dimethylamine oxide molecule as discussed previously (5). More recently, the cardiolipin was identified in a nonfunctioning RC mutant (5). The other two lipids were not found presumably because the mutations caused significant structural changes involving the binding sites of the two lipids.
Functional Implications of the Bound Lipids.
The RC has two branches of cofactors arranged according to an approximate 2-fold symmetry axis. Despite this symmetry, only one branch of the cofactors is active in the electron transfer process. Extensive characterization of modified RCs has led to the general conclusion that the asymmetry of electron transfer is largely due to differences in the energetics of the two possible pathways (21–24). Models have been proposed suggesting a variety of interactions involving the cofactors that could lead to the asymmetry of electron transfer despite the overall structural symmetry; however, alterations of a significant fraction of the amino acid residues surrounding the cofactors have lead to only a limited activation of the second branch (25).
The asymmetric presence of the lipids on the surface suggests that lipid–cofactor interactions may have a significant contribution to the factors that establish the difference in energetics for the two branches of cofactors (Fig. 3). The active bacteriochlorophyll has the nearby tightly bound glycolipid, which shields the cofactor and produces a relatively hydrophobic environment for the cofactor. In contrast, the inactive bacteriochlorophyll has no bound lipid and instead is found to interact with bound solvent molecules that establish polar interactions. The hydrophobic environment should have a minimal response to the reduction of the bacteriochlorophyll monomer, and hence the transfer of an electron from the bacteriochlorophyll dimer to the bacteriopheophytin through the bacteriochlorophyll monomer should have a small reorganization energy and hence be optimized. The possible functional significance of this lipid is supported by the modeling of detergent molecules at a very similar position near the active bacteriochlorophyll monomer in RCs from Blastochloris viridis (formerly Rhodopseudomonas viridis; ref. 26). Also contributing to the asymmetry is the presence of the phosphatidylcholine near only the inactive bacteriopheophytin. The negatively charged phosphate of the lipid is located ≈6.5 Å from the cofactor and may establish an electrostatic interaction that could destabilize the reduced state of the cofactor and make electron transfer to this cofactor energetically unfavorable, compared with the active branch.
Model of Quasiproteins in Membranes.
The lipids were found to be arranged on the surface of the protein with a separation of 30 Å between the polar (or saccharide) groups. In addition to the bacterial RC, a few proteins have now been crystallized that have been found to possess bound lipids (27). Bacteriorhodopsin was found to contain several lipids, although this protein was crystallized by using lipids unlike most other proteins (28). A few lipids have been reported for cytochrome c oxidase (29, 30), FhuA (31), and photosystem I (32). The presence of these lipids after extensive purification shows that they must have significant interactions with the protein surface. By extrapolation, in the cell membrane, the proteins would have a range of interactions with a large number of lipids. Thus, the proteins would not behave as simple independent components but rather as “quasiproteins,” or protein–lipid complexes. The shell of interacting lipids provides the complex with functional properties different from those of the bare isolated protein just as the charges of ions in solution are shielded by clouds of counterions. The presence of interacting lipids surrounding the integral membrane proteins may be critical for certain functions of these proteins in cell membranes. For example, in photosynthetic organisms, there may be specific lipids interacting near QB of the RC that facilitate the release of quinol and insertion of quinone from the surrounding pool. In general, the interactions between proteins and lipids in the complex environment of the cell membrane are currently poorly understood, and the possible roles of lipids in influencing protein function remain an outstanding biochemical question.
Acknowledgments
This work was supported by grants from the National Aeronautics and Space Administration (NAG8-1353) and the National Science Foundation (MCB-0131764).
Abbreviations
RC, reaction center
MALDI, matrix-assisted laser desorption ionization
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code ).
Brune, D. C. & Cheung, J., Proceedings of the 45th ASMS Conference on Mass Spectrometry and Allied Topics, June 1–5, 1997, Palm Springs, CA, p. 853 (abstr.).
References
- 1.Benning C. (1998) in Lipids in Photosynthesis: Structure, Function, and Genetics, eds. Siegenthaler, P. A. & Murata, N. (Kluwer, Dordrecht, The Netherlands), pp. 83–101.
- 2.Michel H., (1991) Crystallization of Membrane Proteins (CRC, Boca Raton, FL).
- 3.Feher G., Allen, J. P., Okamura, M. Y. & Rees, D. C. (1989) Nature (London) 339, 111-116. [Google Scholar]
- 4.Blankenship R. E., Madigan, M. T. & Bauer, C. E., (1995) Anoxygenic Photosynthetic Bacteria (Kluwer, Dordrecht, The Netherlands).
- 5.McAuley K. E., Fyfe, P. K., Ridge, J. P., Isaacs, N. W., Cogdell, R. J. & Jones, M. R. (1999) Proc. Natl. Acad. Sci. USA 96, 14706-14711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams J. C. & Taguchi, A. K. W. (1995) in Anoxygenic Photosynthetic Bacteria, eds. Blankenship, R. E., Madigan, M. T. & Bauer, C. E. (Kluwer, Dordrecht, The Netherlands), pp. 1029–1065.
- 7.Lin X., Murchison, H. A., Nagarajan, V., Parson, W. W., Allen, J. P. & Williams, J. C. (1994) Proc. Natl. Acad. Sci. USA 91, 10265-10269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ermler U., Fritzsch, G., Buchanan, S. K. & Michel, H. (1994) Structure (London) 2, 925-936. [DOI] [PubMed] [Google Scholar]
- 9.Otwinowski Z. & Minor, W. (1997) Methods Enzymol. 276, 307-326. [DOI] [PubMed] [Google Scholar]
- 10.Leslie A. G. W. (1999) Acta Crystallogr. D 55, 1696-1702. [DOI] [PubMed] [Google Scholar]
- 11.CCP4, Collaborative Computing Project Number 4 (1994) Acta Crystallogr. D 50, 760-763.15299374 [Google Scholar]
- 12.Brunger A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 13.Jones T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 14.Laskowski R. A., MacArthur, M. W, Moss, D. S. & Thorton, J. M. (1993) J. Appl. Crystallogr. 26, 283-291. [Google Scholar]
- 15.McRee D. E. & David, P. R., (1999) Practical Protein Crystallography (Academic, New York).
- 16.McCarley T. D., McCarley, R. L. & Limbach, P. A. (1998) Anal. Chem. 70, 4376-4379. [Google Scholar]
- 17.Lancaster C. R. & Michel, H. (1997) Structure (London) 5, 1339-1359. [DOI] [PubMed] [Google Scholar]
- 18.Stowell M. H. B., McPhillips, T. M., Rees, D. C., Soltis, S. M., Abresch, E. & Feher, G. (1997) Science 276, 812-816. [DOI] [PubMed] [Google Scholar]
- 19.Allen J. P., Feher, G., Yeates, T. O., Komiya, H. & Rees, D. C. (1988) Proc. Natl. Acad. Sci. USA 85, 8487-8491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang C. H., El-Kabbani, O., Tiede, D., Norris, J. & Schiffer, M. (1991) Biochemistry 30, 5352-5360. [DOI] [PubMed] [Google Scholar]
- 21.Heller B. A., Holten, D. & Kirmaier, C. (1995) Science 269, 940-945. [DOI] [PubMed] [Google Scholar]
- 22.van Brederode M. E., van Mourik, F., van Stokkum, I. H. M., Jones, M. R. & van Grondelle, R. (1999) Proc. Natl. Acad. Sci. USA 96, 2054-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirmaier C., He, C. & Holten, D. (2001) Biochemistry 40, 12132-12139. [DOI] [PubMed] [Google Scholar]
- 24.Katilius E., Katiliene, Z., Lin, S., Taguchi, A. K. W. & Woodbury, N. W. (2002) J. Phys. Chem. B 1063, 11471-11475. [Google Scholar]
- 25.Woodbury N. W. & Allen, J. P. (1995) in Anoxygenic Photosynthetic Bacteria, eds. Blankenship, R. E., Madigan, M. T. & Bauer, C. E. (Kluwer, Dordrecht, The Netherlands), pp. 527–557.
- 26.Deisenhofer J., Epp, O., Sinning, I. & Michel, H. (1995) J. Mol. Biol. 246, 429-457. [DOI] [PubMed] [Google Scholar]
- 27.Garavito R. M. & Ferguson-Miller, S. (2001) J. Biol. Chem. 276, 22403-22406. [DOI] [PubMed] [Google Scholar]
- 28.Belrhali H., Nollert, P., Royant, A., Menzei, C., Rosenbusch, J. P., Landau, E. M. & Pebay-Peyroula, E. (1999) Structure (London) 7, 909-917. [DOI] [PubMed] [Google Scholar]
- 29.Tsukihara T., Aoyama, H., Yamashita, E., Tomizaki, T., Yamaguchi, H., Shinzawa-Itoh, K., Nakashima, R., Yaono, R. & Yoshikawa, S. (1996) Science 272, 1136-1144. [DOI] [PubMed] [Google Scholar]
- 30.Harrenga A. & Michel, H. (1999) J. Biol. Chem. 274, 33296-33299. [DOI] [PubMed] [Google Scholar]
- 31.Ferguson A. D., Hofmann, E., Coulton, J. W., Diederichs, K. & Welte, W. (1998) Science 282, 2215-2220. [DOI] [PubMed] [Google Scholar]
- 32.Jordon P., Fromme, P., Witt, H. T., Klukas, O., Saenger, W. & Krauss, N. (2001) Nature (London) 411, 909-917. [DOI] [PubMed] [Google Scholar]