Abstract
SOS mutagenesis in Escherichia coli requires DNA polymerase V (pol V) and RecA protein to copy damaged DNA templates. Here we show that two distinct biochemical modes for RecA protein are necessary for pol V-catalyzed translesion synthesis. One RecA mode is characterized by a strong stimulation in nucleotide incorporation either directly opposite a lesion or at undamaged template sites, but by the absence of lesion bypass. A separate RecA mode is necessary for translesion synthesis. The RecA1730 mutant protein, which was identified on the basis of its inability to promote pol V (UmuD′2C)-dependent UV-mutagenesis, appears proficient for the first mode of RecA action but is deficient in the second mode. Data are presented suggesting that the two RecA modes are “nonfilamentous”. That is, contrary to current models for SOS mutagenesis, formation of a RecA nucleoprotein filament may not be required for copying damaged DNA templates. Instead, SOS mutagenesis occurs when pol V interacts with two RecA molecules, first at a 3′ primer end, upstream of a template lesion, where RecA mode 1 stimulates pol V activity, and subsequently at a site immediately downstream of the lesion, where RecA mode 2 cocatalyzes lesion bypass. We posit that in vivo assembly of a RecA nucleoprotein filament may be required principally to target pol V to a site of DNA damage and to stabilize the pol V-RecA interaction at the lesion. However, it is only a RecA molecule located at the 3′ filament tip, proximal to a damaged template base, that is directly responsible for translesion synthesis.
Keywords: SOS mutagenesis, error-prone DNA polymerase, DNA damage-induced mutation
The ability to survive extensive DNA damage depends on the presence of DNA repair pathways including base excision repair, nucleotide excision repair, and postreplication mismatch repair, all of which seem to be conserved to a remarkable degree spanning the simplest unicellular to the most complex higher organisms (1). However, despite the high efficiency of DNA repair, replication forks are likely to encounter impenetrable “road blocks” in the form of persisting template lesions. In Escherichia coli, umuC and umuD (2) are among the more than 40 genes induced to cope with DNA damage as part of the SOS regulon (3). UmuC and UmuD′ (the shorter, mutagenically active form of UmuD) combine as a heterotrimer (4, 5) to form an error-prone DNA polymerase, pol V (UmuD′2C) (6–8), that substitutes for the replicative pol III core enabling replication of a damaged template strand.
Pol V is the major translesion synthesis (TLS) polymerase in E. coli (9). In contrast to the other two SOS-induced polymerases, pol II and pol IV, which are induced rapidly after DNA damage (10, 11), pol V is virtually undetectable until about 20–40 min after SOS induction (12–14). The delayed expression of pol V provides an opportunity for a cell to repair its damaged DNA before being forced into undertaking potentially mutagenic translesion replication.
The successful reconstitution of an in vitro assay measuring pol V-catalyzed TLS has provided an opportunity to investigate the biochemical mechanisms governing error-prone DNA replication (6, 15, 16). Three protein components were previously found to be required for TLS: pol V, RecA, and E. coli single-stranded binding protein (SSB) (6). TLS was stimulated in the presence of the β sliding clamp and γ clamp-loading complex (β/γ), but the processivity proteins are not an absolute requirement in vitro (17, 18).
The key to understanding the mechanism of pol V-catalyzed TLS lies in discerning the precise role(s) of RecA protein. RecA is known to play three independent roles in the cell (19). By forming a RecA nucleoprotein filament on single-stranded DNA (ssDNA), RecA catalyzes DNA strand exchange during homologous recombination (20–23). A RecA nucleoprotein filament is also required to induce the SOS response by serving as a coprotease to stimulate LexA's latent capacity to autodigest (24) thereby inactivating its repressor function (24–26). In an analogous reaction, UmuD is cleaved generating mutagenically active UmuD′ (27–29), which then interacts with UmuC to form pol V (4–6).
Compelling evidence for a direct role of RecA in SOS mutagenesis was obtained through the genetic characterization of various recA mutants (29–31). Of particular interest is recA1730 (S117F) (30). This RecA mutant is unable to promote UmuD′2C-dependent UV-induced mutagenesis, but is proficient for mutagenesis in the presence of the related MucA′B proteins (32) and also retains some proficiency in both recombination and SOS induction when expressed at sufficiently high levels (33).
Here we address the biochemical mechanism governing the roles of RecA protein during pol V-catalyzed TLS. Because the RecA nucleoprotein filament is the active form of RecA in its recombination and coprotease functions, it has been assumed that RecA filaments would be needed for SOS mutagenesis. A previous study suggested that a filament is required for TLS (34). Here, we address two issues. The first is to determine the biochemical roles of RecA protein during TLS. The second concerns whether or not the formation of a RecA nucleoprotein filament per se is a prerequisite to achieve lesion bypass. We also discuss the roles of RecA and pol V in relation to the roles of SSB and β/γ complex (18).
Materials and Methods
Adenosine 5′-[γ-thio]triphosphate (ATP-γ-S) was purchased from Roche Molecular Biochemicals. Proteins and other chemicals were described (17). Templates used in this study were 120-mer and 36-mer synthetic oligonucleotides with or without a synthetic abasic (tetrahydrofuran) lesion. 5′-32P-labeled or unlabeled 30-mers were used as primers. Pol V (UmuD′2C) was purified as a native, untagged protein (5, 7) from a strain carrying deletion mutations in polB and dinB, encoding DNA polymerases II and IV, respectively. Wild-type RecA and mutant RecA1730 proteins were purified as described (21).
Nucleotide Incorporation on Lesion-Containing and Natural DNA Primer Templates.
All reaction mixtures (10 μl) contained 20 mM Tris (pH 7.5), 8 mM MgCl2, 5 mM DTT, 0.1 mM EDTA, 25 mM sodium glutamate, 40 μg ml−1 BSA, and 4% (vol/vol) glycerol. Reactions were performed at 37°C in the presence of 30-mer/36-mer primer-template (p/t) DNA (10 nM or 360 nM template nt), pol V (240 nM), ATP-γ-S (0.5 mM), RecA protein (varied as indicated in the figures), and the four dNTP substrates (200 μM each). Similar reactions were performed by using a primed 120-mer template containing 2- and 3-nt gaps or without gaps, except that SSB (250 nM) was present in some reactions, as indicated in the figures. The reactions were conducted at 37°C for 10 min and terminated by adding 20 μl of 20 mM EDTA in 95% formamide. Synthesis products were heat-denatured and separated on a 12% polyacrylamide denaturing gel; band intensities were measured by phosphorimaging with IMAGEQUANT software (Molecular Dynamics). Primer template utilization was computed from the integrated gel band intensities as the fraction of extended primers in the reaction. The TLS efficiency was calculated as the ratio of primers extended past the lesion divided by the total number of extended primers.
RecA Protein Binding to DNA Modified with 2-Aminopurine (2AP).
2AP is located at the 5′ end of the template strand. Reactions (70 μl) contained 20 mM Tris⋅HCl (pH 7.5), 8 mM MgCl2, 5 mM DTT, 25 mM sodium glutamate, 0.5 mM ATP-γ-S, 10 μM DNA template nt, and increasing amounts of RecA or RecA1730 protein. The binding assays were performed at 37°C, for 3 min at each concentration of RecA, and fluorescence was measured by using a PTI (Photon Technology International, Lawrenceville, NJ) QuantaMaster 1 fluorimeter at an excitation wavelength of 310 nm and an emission wavelength of 362 nm, with a 3-nm bandpass width. Relative fluorescence increase (RFI) was calculated by determining the RecA protein intrinsic fluorescence, and by dividing the RecA + DNA fluorescence intensity by the fluorescence intensity of DNA alone.
Results
The principle goal of this study is to determine the role of RecA protein during pol V-catalyzed TLS. The active form of the RecA protein is a nucleoprotein filament during homologous recombination and SOS induction (35). Because RecA protein does not act “alone,” either as a monomer or small multimer in these two prototypic reactions, one might assume that a nucleoprotein filament is also required to perform SOS-induced TLS (34). We address two mechanistic questions in this paper. Is formation of a RecA nucleoprotein filament required per se for TLS? What are the RecA–pol V interactions required to copy past damaged DNA template bases?
Pol V-Catalyzed TLS on Short ssDNA Template Overhangs.
We have previously observed maximum levels of TLS, when copying 30-mer/120-mer p/t DNA, in the presence of 1 RecA protein to about 5 nt DNA (18), which is similar to the stoichiometry of RecA protein binding to ssDNA (1 RecA:3 DNA template nt) (23, 35). Unexpectedly, we also observed significant TLS (nearly half the maximum level) with 1 RecA per 50 template nt (18), going as low as 1 RecA per 120 nt (data not shown). Filament formation is not likely at such small RecA/DNA nt ratios on these short p/t DNA substrates, even when making allowances for the cooperative, inhomogeneous nature of nucleoprotein filament assembly (23, 35). These data suggested that although RecA is absolutely required for TLS, the assembly of a RecA nucleoprotein filament may not be required. However, we could not exclude the possibility that TLS occurred on a minority of p/t DNA having a RecA filament present downstream of the 3′ primer end.
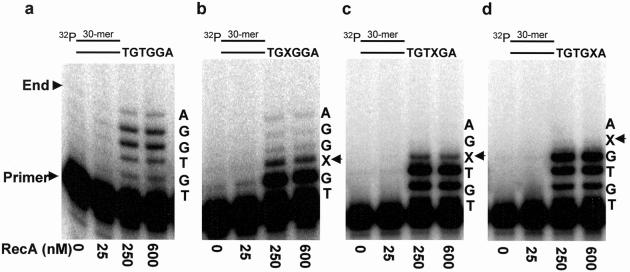
To assess whether or not a RecA filament is necessary for TLS, we measured pol V synthesis on 30-mer/36-mer p/t DNA containing a short 5′ template overhang. The TLS data are illustrated with a 6-nt template overhang containing a lesion located adjacent to either 1, 2, or 3 normal nt from the 5′ template end (Fig. 1). This p/t DNA construct should preclude nucleoprotein filament assembly because only enough space exists to bind two RecA molecules ahead of the 3′ primer end.
Fig 1.
Pol V-catalyzed DNA synthesis as a function of RecA concentration on a 6-nt DNA template overhang, depicting two modes of nonfilamentous RecA action. (a) Undamaged DNA template; (b) template abasic lesion located adjacent to 3 normal nt from the 5′ template end; (c) template abasic lesion located adjacent to 2 normal nt from the 5′ template end; (d) template abasic lesion located adjacent to 1 normal nt from the 5′ template end. Arrows indicate the location of an abasic template lesion, X. The p/t DNA is shown at the top of each panel. The p/t DNA concentration was 10 nM. The ratio of RecA:DNA nt for each data point is: 25 nM RecA (1 RecA:15 DNA template nt), 250 nM (1:1.5), 600 nM RecA (1:0.6). The reactions were performed in the absence of SSB.
The control template without a lesion is copied completely by the addition of 6 nt, in a reaction strongly stimulated at increasing concentrations of RecA protein (Fig. 1a). Synthesis to the end of the template occurs also when the lesion is located 4 nt from the 5′ end of the template overhang, leaving a region of three undamaged template bases sufficient for binding a single RecA monomer (Fig. 1b). Synthesis occurs directly opposite the lesion but proceeds no further when two undamaged template bases are present downstream from the lesion (Fig. 1c), and no synthesis occurs opposite X when just one downstream template base is present (Fig. 1d).
Two Distinct RecA-Binding Modes Are Required for Pol V-Catalyzed TLS.
The strong stimulation in nucleotide incorporation caused by RecA before encountering the lesion, whether the lesion is bypassed (Fig. 1b) or not (Fig. 1 c and d), suggests that RecA is likely to be playing at least two different roles during TLS. In mode 1, RecA stimulates pol V, perhaps by direct interaction with pol V, and permits DNA synthesis up to and opposite the DNA lesion, but not beyond it (Fig. 1 c and d). Mode 2 is required for lesion bypass and involves interaction of RecA with a short segment of DNA (≥3 nt) beyond the lesion (Fig. 1b).
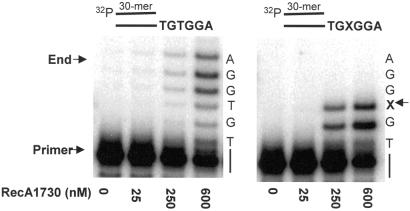
We tested this “two mode” idea by using a mutant RecA, RecA1730 (S117F), to determine its effect on pol V-catalyzed TLS with a 6-nt template overhang containing a region of three undamaged bases downstream from the lesion. The RecA1730 mutant is defective in pol V (Umu)-dependent mutagenesis, but is proficient for MucA′B-dependent mutagenesis as well as retaining some capacity to promote homologous recombination and induction of SOS when overexpressed (12, 30, 32, 33). Pol V-catalyzed TLS is almost abolished in the presence of RecA1730 (Fig. 2b) despite the fact that incorporation occurs on both undamaged DNA and directly opposite the lesion (Fig. 2). The fraction of primers extended opposite X that continue past the lesion is less than 0.3% despite having extension of about 33% of the primer strands (Fig. 2 Right, lane 4). In contrast, the TLS fraction for wild-type RecA is about 3% when only 8% of the primers are extended (Fig. 1b, lanes 3 and 4). These data reinforce the idea that wild-type RecA plays two distinct roles: (i) a stimulatory effect on pol V synthesis at both damaged and undamaged template sites, and (ii) as a required cofactor enabling pol V to copy past a lesion, which is essentially absent with RecA1730.
Fig 2.
Pol V-catalyzed DNA synthesis as a function of mutant RecA1730 (S117F) concentration on a short DNA template overhang, depicting the presence of mode 1 RecA stimulation of pol V activity and the absence of mode 2 RecA cocatalyzed translesion synthesis. (Left) Synthesis on an undamaged 6-nt DNA template overhang. (Right) Synthesis on a 6-nt DNA template overhang containing an abasic lesion, X, located 4 nt from the 5′ template end. The p/t DNA concentration was 10 nM. The ratio of RecA/DNA nt for each data point was: 25 nM RecA (1 RecA:15 DNA template nt), 250 nM (1:1.5), 600 nM RecA (1:0.6). The reactions were performed in the absence of SSB.
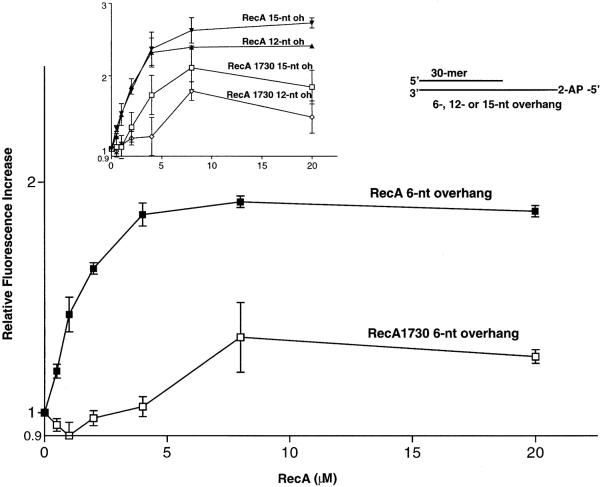
RecA protein binds to ssDNA in a stoichiometric manner (1 RecA monomer per 3 template nt) by forming a right-handed helical filament (35). However, our data demonstrate that pol V-catalyzed TLS depends on the presence of RecA even when as few as 3 nt are present downstream of a lesion, which is barely sufficient to bind a single RecA monomer. We examined the ability of RecA protein to bind in the absence of pol V to the same p/t DNA used in the lesion bypass assay, but with 2AP present at the 5′ template end. An increase in 2AP fluorescence intensity is a measure of the steady-state level of RecA bound to p/t DNA.
Wild-type RecA protein in the presence of ATP-γ-S interacts more strongly with p/t DNA containing a 6-nt template overhang compared with RecA1730, as determined by a greater increase in 2AP fluorescence intensity (Fig. 3). Binding to p/t DNA is also greater for wild type compared with mutant RecA in the presence of ATP, but the binding seems much weaker on the basis of about a 10-fold smaller increase in relative fluorescence (data not shown). Binding of wild-type RecA to p/t DNA increases with the length of the ssDNA overhang (Fig. 3 Inset). In each case, the wild-type RecA binds more avidly to the p/t DNA than does RecA1730. The 2AP fluorescence-binding data were confirmed independently by measuring rates of DNA-dependent RecA-catalyzed ATP hydrolysis as a function of the length of the ssDNA overhang. The hydrolysis rates with wild-type RecA were 0.04 min−1 and 1 min−1 for the 6- and 15-nt template overhangs, respectively. These are significantly reduced compared with ATP hydrolysis rates (≈30 min−1) on long ssDNA (data not shown) (36).
Fig 3.
Binding of RecA protein to primer-template DNA containing short ssDNA template overhangs. Binding by RecA and RecA1730 proteins was detected as an increase in fluorescence intensity of 2AP located on the 5′ end of the template strand. The DNA concentration in each reaction was 10 μM nt. The two curves depict wild-type RecA protein binding to p/t DNA with a 6-nt overhang (oh) and RecA1730 protein binding to p/t DNA with a 6-nt oh. (Inset) The data depict wild-type RecA binding to p/t DNA with a 15-nt oh; RecA protein binding to p/t DNA with a 12-nt oh; RecA 1730 protein binding to p/t with a 15-nt oh; RecA1730 protein binding to p/t with a 12-nt oh. The DNA concentration in each reaction was 10 μM nt. The ratio of RecA/DNA nt for each data point was: 0.5 μM RecA (1 RecA:20 DNA template nt), 1 μM RecA (1:10), 2 μM RecA (1:5), 4 μM RecA (1:2.5), 8 μM RecA (1:1.25), 20 μM RecA (2:1). The p/t DNA is shown in the sketch at the top.
Pol V-Catalyzed TLS on 3- and 2-Nucleotide Internal Gaps.
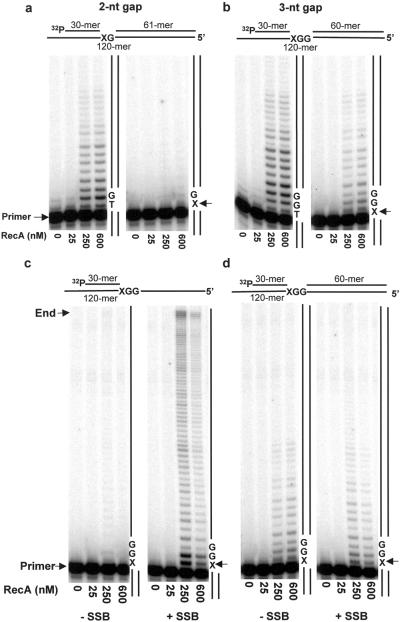
Pol V-catalyzed TLS requires at least 3 template nt downstream from a lesion when copying a variety of template overhangs, as illustrated for a 30-mer/36-mer p/t DNA containing a 6 nt ssDNA 5′-overhang (Fig. 1). The data are consistent with having 3 nt bound by 1 RecA molecule (35). To investigate this point further, we measured the ability of pol V to copy short internal ssDNA gaps containing either undamaged or damaged template bases.
Increased synthesis occurs with increasing RecA levels within a 3-nt gap containing undamaged DNA (Fig. 4b Left). When copying a damaged template strand, TLS occurs readily in the 3-nt gap at 250 nM and 600 nM RecA (Fig. 4b Right). However, TLS is essentially absent in the 2-nt gap (Fig. 4a Right) even though the 2-nt gap is filled when no lesion is present (Fig. 4a Left). Because the “footprint” of a single RecA monomer is 3 nt DNA, the observation that TLS can occur within a 3-nt but not a 2-nt gap is again consistent with the need to bind at least a single RecA protein monomer for lesion bypass.
Fig 4.
Short-gap-filling reactions catalyzed by pol V as a function of RecA concentration in the presence and absence of a template abasic lesion. (a) 2-nt DNA template gap; (b) 3-nt DNA template gap; (c) 63-nt ssDNA template overhang, containing an abasic lesion, in the absence and presence of SSB; (d) 3-nt DNA template gap, containing an abasic lesion, in the absence and presence of SSB. ATP-γ-S was present in all reactions. The p/t DNA concentration was 10 nM. The ratio of RecA/DNA nt for each data point was: 25 nM RecA (1 RecA:50 DNA template nt); 250 nM RecA (1:5); 600 nM RecA (1:2). The arrows indicate the location of an abasic template lesion, X. The p/t DNA is shown at the top of each panel.
A decided stimulation of pol V synthesis by RecA also occurs in both gaps when no lesion is present. Because room in the 2-nt gap is insufficient to bind a RecA molecule, the data reinforce the existence of an independent mode of RecA action, one which stimulates pol V activity on undamaged DNA, but does not influence lesion bypass.
The synthesis taking place beyond the minimum gap-filling reaction (Fig. 4 a and b) is caused by strand displacement rather than by the absence of double-stranded (ds) DNA downstream from the lesion. This point is verified by the virtual elimination of TLS on the ssDNA template downstream from the lesion in the absence of a gap (i.e., where no dsDNA is present downstream of X) (Fig. 4c Left). In contrast, TLS occurs within the gapped template regions (Fig. 4b Right) and on the short template overhang having at least 3 nt present downstream from the lesion (Fig. 1b).
The key point is that the assembly of a RecA nucleoprotein filament on a long ssDNA template actually blocks pol V synthesis (18) (Fig. 4c Left). The block to synthesis is alleviated by the presence of SSB, which acts in concert with pol V to disassemble the RecA filament in a 3′ to 5′ direction, ahead of the advancing pol V, akin to a “locomotive cowcatcher” (18) (Fig. 4c Right). In contrast to the requirement for SSB to copy RecA-coated DNA, SSB has essentially no effect on pol V-catalyzed TLS within short gapped regions (Fig. 4d). Thus, the data using pol V + RecA to copy the short gapped p/t DNA substrates provide additional support for the two main points of the paper: (i) assembly of a RecA nucleoprotein filament is not a requirement for TLS; (ii) RecA has two distinct modes of action during TLS.
Discussion
RecA protein plays an essential role in disparate biochemical pathways in E. coli. RecA catalyzes DNA strand exchange during homologous recombination (35, 36), acts as a coprotease during cleavage of LexA and UmuD proteins required for SOS induction and pol V assembly (26, 28, 37), and is a required cofactor during SOS-induced mutagenesis (29–31). RecA's roles in homologous recombination and SOS induction are well understood (1). In contrast, its biochemical role in pol V-catalyzed TLS is largely unknown. Because the recombination and cleavage reactions involving RecA require formation of a nucleoprotein filament, it seemed reasonable to presume that a RecA nucleoprotein filament must also be required for TLS. Our data challenge this commonly held perception.
Pol V-Catalyzed TLS Occurs in the Absence of a RecA Nucleoprotein Filament.
The conclusion that a RecA nucleoprotein filament is unnecessary for TLS is based on three lines of evidence. First, we have observed that pol V-catalyzed TLS occurs on short ssDNA overhangs, but only when at least 3 nt are present on the template strand downstream from the lesion (Fig. 1). Second, bypass of an abasic lesion takes place in a 3-nt gap, which is large enough to bind one RecA monomer (Fig. 4b). In contrast, TLS is essentially absent in a 2-nt gap, which is insufficient to bind a RecA monomer (Fig. 4a). A 2-nt gap is filled in the absence of a lesion suggesting that its inability to support TLS relates to the absence of RecA binding in the gap, not to a catalytic deficiency attributable to pol V.
One might suggest that a RecA filament could still exist, because a RecA monomer bound in the short overhang or gap could be extended to reach the 3′ primer end and into the duplex by normal 5′ to 3′ RecA filament growth. The third line of evidence addresses this possibility by showing that TLS is virtually eliminated when RecA filament assembly takes place on a p/t DNA containing a 63-nt-ssDNA overhang in the absence of SSB (Fig. 4c Left). SSB acting in concert with pol V disassembles the nucleoprotein filament in a 3′ to 5′ direction, ahead of the advancing pol V, enabling TLS to occur (18) (Fig. 4c Right). In contrast, SSB has no discernible effect on TLS in a 3-nt gap (Fig. 4d). We suggest that, in the absence of SSB, occlusion of the 3′ primer end by RecA filaments serves to block pol V access, thereby inhibiting synthesis on damaged and undamaged DNA. This inhibitory effect of RecA occurs on ssDNA tails as short as 15 nt (data not shown), but does not occur on p/t DNA containing 6-nt ssDNA tails.
TLS may be blocked even in the presence of SSB when the concentration of RecA protein is increased so that binding is saturated. Note, for example, the decreased reaction in Fig. 4c when the RecA concentration is increased from 250 nM (1 RecA per 5 template nt) to 600 nM (1 per 2 nt). Higher RecA levels block the reaction almost completely (data not shown). Because binding of a RecA filament across the primer terminus is inhibitory, and no room for filament assembly exists downstream of the 3′ primer end in these substrates, assembly of a RecA nucleoprotein filament is unlikely to be responsible for pol V-catalyzed TLS on 6-nt ssDNA overhangs (Fig. 1).
RecA Has Two Essential Nonfilamentous Roles During Pol V-Catalyzed TLS.
TLS occurs in 3-nt gaps (Fig. 4b) and on 6-nt template overhangs when 3 nt are present downstream from a lesion (Fig. 1b). However, TLS does not occur in 2-nt gaps (Fig. 4a) or when <3 nt are present downstream from a template lesion on short ssDNA template overhangs (Fig. 4 c and d), yet synthesis by pol V is strongly stimulated in the presence of wild-type RecA protein (Figs. 1 and 4). Even when wild-type RecA is unable to bind either in a 2-nt gap or when fewer than 3 nt are present downstream from lesion on a template overhang, RecA is nevertheless able to stimulate pol V activity (Figs. 4b and 1 c and d, respectively).
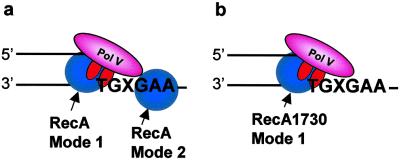
A further demonstration of RecA stimulation of pol V in the absence of TLS is observed in reactions by using the mutant RecA1730 (Fig. 2). E. coli recA1730 mutant strains are deficient in Umu-dependent mutagenesis despite the ability of the mutant protein to catalyze homologous recombination and to induce the SOS response when RecA1730 is overexpressed (29–33). The dependence of pol V activity on RecA1730 coupled with the almost complete absence of RecA1730-dependent TLS, even though 3 nt are present downstream of the lesion (Fig. 2 Right), reinforces the distinct nature of the two wild-type RecA modes of action. Pol V activity is strongly stimulated by RecA mode 1 action, and pol V-catalyzed TLS requires RecA mode 2 action. A model illustrating the two RecA modes (Fig. 5) shows RecA protein interacting with pol V at the 3′ primer end (mode 1) and with DNA and pol V at a template lesion (mode 2). In the case of wild-type RecA, the TLS efficiency is 3% when 8% of the primers are elongated (Fig. 1b), whereas with RecA1730, the TLS efficiency is <0.3% when ≈33% of the primers are elongated (Fig. 2 Right).
Fig 5.
Model depicting pol V-catalyzed SOS translesion synthesis. (a) Wild-type RecA exhibits two modes of RecA action, mode 1 stimulation of pol V activity when interacting with pol V at the 3′-OH primer end, and mode 2 cocatalysis of translesion synthesis when bound to the DNA template strand at the 5′ side of the lesion, X. (b) Mutant RecA1730 (S117F) exhibits mode 1 stimulation of pol V activity but is essentially unable to cocatalyze translesion synthesis, based on the data in Fig. 2. RecA protein is illustrated as a monomer, but its detailed structure is unknown and could instead be a multimer.
Reconciling Filamentous and Nonfilamentous RecA Action During Pol V-Catalyzed TLS.
A RecA nucleoprotein filament is likely to assemble in vivo on a region of ssDNA downstream of a stalled replication fork proximal to a damaged DNA template base. The RecA nucleoprotein filament might then function in two distinct ways, either during recombination-dependent error-free replication restart (38–40), requiring RecFOR, PriA DnaG (41, 42), and DNA pol II (43, 44), or alternatively for recombination-independent error-prone TLS, requiring pol V and SSB (18). In the latter process, assembly of a filament may provide a means of targeting pol V to the lesion site (18, 45). On the basis of the data presented in this paper, only the RecA monomer at the 3′ tip of the nucleoprotein filament is absolutely required for TLS, an idea that had been suggested by Devoret and colleagues (46, 47). The remainder of the RecA filament is likely to have an important role in vivo by stabilizing the 3′-RecA molecule at the lesion site to “cocatalyze” TLS along with pol V and SSB (18).
The presence of SSB has little discernible effect on TLS in the absence of RecA filament assembly when using p/t DNA containing either small gaps or short template overhangs (see, for example, Fig. 4d). In contrast, SSB must be present to copy longer (63 nt) ssDNA overhangs on which a RecA filament has assembled (18) (Fig. 4c). We have shown that pol V + SSB act akin to a locomotive “cowcatcher” to disassemble such a RecA nucleoprotein filament in a 3′ to 5′ direction ahead of an advancing pol V (18). Although our data demonstrate that SSB is not essential for the TLS reaction per se, it could be essential in vivo where RecA nucleoprotein filaments are ubiquitous.
In summary, we have presented evidence that two RecA modes are required for pol V-catalyzed TLS, and that neither mode seems to require assembly of a nucleoprotein filament. RecA mode 1 strongly stimulates pol V activity on both undamaged and damaged DNA, but cannot catalyze TLS. If a nucleoprotein filament is present, then a RecA monomer at the 3′ tip of the filament operates in mode 2 to catalyze lesion bypass. We are cognizant of the uncertainty surrounding the precise form(s) of RecA protein in the two nonfilamentous binding modes. We do not know of any biochemical precedent with RecA acting as a monomer. Indeed, a hexameric form of RecA has been reported in E. coli (48, 49) and Thermophilus aquaticus (50), as well as octameric forms of human Dmc1 (51) and archaeal RadA (52, 53). In light of these multimeric forms of RecA-like proteins, our use of the term RecA “molecule” or “monomer” to designate the two putative nonfilamentous modes (Fig. 5) is used solely as an “Occam's Razor” assumption in lieu of structural data.
Acknowledgments
This work was supported by National Institutes of Health (NIH) Grants GM42554, GM21422 (to M.F.G.) and GM52725 (to M.M.C.), and by the NIH Intramural Research Program (to R.W.). P.P. and E.M.S. were supported by NIH/National Institute on Aging Training Grant AG00093.
Abbreviations
TLS, translesion synthesis
pol, DNA polymerase
SSB, E. coli single-stranded binding protein
p/t DNA, primer-template DNA
ssDNA, single-stranded DNA
dsDNA, double-stranded DNA
2AP, 2-aminopurine
X, an abasic (tetrahydrofuran) moiety
ATP-γ-S, adenosine 5′-[γ-thio]triphosphate
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Friedberg E. C., Walker, G. C. & Siede, W., (1995) DNA Repair and Mutagenesis (Am. Soc. Microbiol. Press, Washington, DC).
- 2.Bagg A., Kenyon, C. J. & Walker, G. C. (1981) Proc. Natl. Acad. Sci. USA 78, 5749-5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Courcelle J. A., Khodursky, A., Peter, B., Brown, P. O. & Hanawalt, P. C. (2001) Genetics 158, 41-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woodgate R., Rajagopalan, M., Lu, C. & Echols, H. (1989) Proc. Natl. Acad. Sci. USA 86, 7301-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruck I., Woodgate, R., McEntee, K. & Goodman, M. F. (1996) J. Biol. Chem. 271, 10767-10774. [DOI] [PubMed] [Google Scholar]
- 6.Tang M., Bruck, I., Eritja, R., Turner, J., Frank, E. G., Woodgate, R., O'Donnell, M. & Goodman, M. F. (1998) Proc. Natl. Acad. Sci. USA 95, 9755-9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang M., Shen, X., Frank, E. G., O'Donnell, M., Woodgate, R. & Goodman, M. F. (1999) Proc. Natl. Acad. Sci. USA 96, 8919-8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reuven N. B., Arad, G., Maor-Shoshani, A. & Livneh, Z. (1999) J. Biol. Chem. 274, 31763-31766. [DOI] [PubMed] [Google Scholar]
- 9.Goodman M. F. (2002) Annu. Rev. Biochem. 70, 17-50. [DOI] [PubMed] [Google Scholar]
- 10.Qiu Z. & Goodman, M. F. (1997) J. Biol. Chem. 272, 8611-8617. [DOI] [PubMed] [Google Scholar]
- 11.Kim S., Matsui, K., Yamada, M., Gruz, P. & Nohmi, T. (2001) Mol. Genet. Genomics 266, 207-215. [DOI] [PubMed] [Google Scholar]
- 12.Sommer S., Boudsocq, F., Devoret, R. & Bailone, A. (1998) Mol. Microbiol. 28, 281-291. [DOI] [PubMed] [Google Scholar]
- 13.Opperman T., Murli, S., Smith, B. T. & Walker, G. C. (1999) Proc. Natl. Acad. Sci. USA 96, 9218-9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodman M. F. & Woodgate, R. (2000) Cold Spring Harbor Symp. Quant. Biol. 65, 31-40. [DOI] [PubMed] [Google Scholar]
- 15.Rajagopalan M., Lu, C., Woodgate, R., O'Donnell, M., Goodman, M. F. & Echols, H. (1992) Proc. Natl. Acad. Sci. USA 89, 10777-10781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reuven N. B., Tomer, G. & Livneh, Z. (1998) Mol. Cell 2, 191-199. [DOI] [PubMed] [Google Scholar]
- 17.Tang M., Pham, P., Shen, X., Taylor, J.-S., O'Donnell, M., Woodgate, R. & Goodman, M. F. (2000) Nature (London) 404, 1014-1018. [DOI] [PubMed] [Google Scholar]
- 18.Pham P., Bertram, J. G., O'Donnell, M., Woodgate, R. & Goodman, M. F. (2001) Nature (London) 409, 366-370. [DOI] [PubMed] [Google Scholar]
- 19.Echols H. & Goodman, M. F. (1990) Mutat. Res. 236, 301-311. [DOI] [PubMed] [Google Scholar]
- 20.Das Gupta C., Shibata, T., Cunningham, R. P. & Radding, C. M. (1980) Cell 22, 437-446. [DOI] [PubMed] [Google Scholar]
- 21.Cox M. M. & Lehman, I. R. (1981) Proc. Natl. Acad. Sci. USA 78, 3433-3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Radding C. M. (1991) J. Biol. Chem. 266, 5355-5358. [PubMed] [Google Scholar]
- 23.Lusetti S. L. & Cox, M. M. (2002) Annu. Rev. Biochem. 71, 71-100. [DOI] [PubMed] [Google Scholar]
- 24.Little J. W. (1984) Proc. Natl. Acad. Sci. USA 81, 1375-1379.6231641 [Google Scholar]
- 25.Little J. W., Edmiston, S. H., Pacelli, L. Z. & Mount, D. W. (1980) Proc. Natl. Acad. Sci. USA 77, 3225-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Little J. W. & Mount, D. W. (1982) Cell 29, 11-22. [DOI] [PubMed] [Google Scholar]
- 27.Shinagawa H., Iwasaki, T., Kato, T. & Nakata, A. (1988) Proc. Natl. Acad. Sci. USA 85, 1806-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burckhardt S. E., Woodgate, R., Scheuremann, R. H. & Echols, H. (1988) Proc. Natl. Acad. Sci. USA 85, 1811-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nohmi T., Battista, J. R., Dodson, L. A. & Walker, G. C. (1988) Proc. Natl. Acad. Sci. USA 85, 1816-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dutreix M., Moreau, P. L., Bailone, A., Galibert, F., Battista, J. R., Walker, G. C. & Devoret, R. (1989) J. Bacteriol. 171, 2415-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sweasy J. B., Witkin, E. M., Sinha, N. & Roegner-Maniscalco, V. (1990) J. Bacteriol. 172, 3030-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bailone A., Sommer, S., Knezevic, J., Dutreix, M. & Devoret, R. (1991) Biochemie 73, 479-484. [DOI] [PubMed] [Google Scholar]
- 33.Dutreix M., Burnett, B., Bailone, A., Radding, C. M. & Devoret, R. (1992) Mol. Gen. Genet. 232, 489-497. [DOI] [PubMed] [Google Scholar]
- 34.Reuven N. B., Arad, G., Stasiak, A. Z., Stasiak, A. & Livneh, Z. (2001) J. Biol. Chem. 276, 5511-5517. [DOI] [PubMed] [Google Scholar]
- 35.Kuzminov A. (1999) Microbiol. Mol. Biol. Rev. 63, 751-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kowalczykowski S. C., Dixon, D. A., Eggleston, A. K., Lauder, S. D. & Rehrauer, W. M. (1994) Microbiol. Rev. 58, 401-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walker G. C. (1984) Microbiol. Rev. 48, 60-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khidhir A. M., Casaregola, S. & Holland, I. B. (1985) Mol. Gen. Genet. 199, 133-140. [DOI] [PubMed] [Google Scholar]
- 39.Witkin E. M., Maniscalco, R. V., Sweasy, J. B. & McCall, J. O. (1987) Proc. Natl. Acad. Sci. USA 84, 6805-6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sweasy J. B. & Witkin, E. M. (1991) Biochemie 73, 437-448. [DOI] [PubMed] [Google Scholar]
- 41.Michel B. (2000) Trends Biochem. Sci. 25, 173-178. [DOI] [PubMed] [Google Scholar]
- 42.Marians K. J. (2000) Trends Biochem. Sci. 25, 185-189. [DOI] [PubMed] [Google Scholar]
- 43.Rangarajan S., Woodgate, R. & Goodman, M. F. (1999) Proc. Natl. Acad. Sci. USA 96, 9224-9229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rangarajan S., Woodgate, R. & Goodman, M. F. (2002) Mol. Microbiol. 43, 617-628. [DOI] [PubMed] [Google Scholar]
- 45.Goodman M. F. (2000) Trends Biochem. Sci. 25, 189-195. [DOI] [PubMed] [Google Scholar]
- 46.Sommer S., Bailone, A. & Devoret, R. (1993) Mol. Microbiol. 10, 963-971. [DOI] [PubMed] [Google Scholar]
- 47.Boudsocq F., Campbell, M., Devoret, R. & Bailone, A. (1997) J. Mol. Biol. 270, 201-211. [DOI] [PubMed] [Google Scholar]
- 48.Heuser J. & Griffith, J. (1989) J. Mol. Biol. 210, 473-484. [DOI] [PubMed] [Google Scholar]
- 49.Logan K. M., Skiba, M. C., Eldin, S. & Knight, K. L. (1997) J. Mol. Biol. 266, 306-316. [DOI] [PubMed] [Google Scholar]
- 50.Yu X. & Egelman, E. H. (1997) Nat. Struct. Biol. 4, 101-104. [DOI] [PubMed] [Google Scholar]
- 51.Passy S. I., Yu, X., Li, Z., Radding, C. M., Masson, J.-Y., West, S. C. & Egelman, E. H. (1999) Proc. Natl. Acad. Sci. USA 96, 10684-10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang S., Yu, X., Seitz, E. M., Kowalczykowski, S. C. & Egelman, E. H. (2001) J. Mol. Biol. 314, 1077-1085. [DOI] [PubMed] [Google Scholar]
- 53.McIlwraith M. J., Hall, D. R., Stasiak, A. Z., Stasiak, A., Wigley, D. B. & West, S. C. (2001) Nucleic Acids Res. 29, 4509-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]