Abstract
The serine proteases sequentially activated to form a fibrin clot are inhibited primarily by members of the serpin family, which use a unique β-sheet expansion mechanism to trap and destroy their targets. Since the discovery that serpins were a family of serine protease inhibitors there has been controversy as to the role of conformational change in their mechanism. It now is clear that protease inhibition depends entirely on rapid serpin β-sheet expansion after proteolytic attack. The regulatory advantage afforded by the conformational mobility of serpins is demonstrated here by the structures of native and S195A thrombin-complexed heparin cofactor II (HCII). HCII inhibits thrombin, the final protease of the coagulation cascade, in a glycosaminoglycan-dependent manner that involves the release of a sequestered hirudin-like N-terminal tail for interaction with thrombin. The native structure of HCII resembles that of native antithrombin and suggests an alternative mechanism of allosteric activation, whereas the structure of the S195A thrombin–HCII complex defines the molecular basis of allostery. Together, these structures reveal a multistep allosteric mechanism that relies on sequential contraction and expansion of the central β-sheet of HCII.
The predominant protease inhibitors of the higher organisms, the serpins (1, 2), have evolved a complex mechanism that was revealed recently by the crystallographic structure of a serpin–protease complex (3). This dramatic mechanism amounts to a race between proteolysis and the incorporation of the serpin-reactive center loop into its main β-sheet (β-sheet A). The dependence of the serpin inhibitory mechanism on β-sheet expansion renders serpins highly susceptible to both loss-of-function and gain-of-function diseases but additionally allows for a range of mechanisms for the modulation of serpin activity. Thus, serpins typically are found controlling tightly regulated physiological pathways such as blood coagulation. Antithrombin (AT) and heparin cofactor II (HCII) have independently (4, 5) evolved mechanisms by which they circulate in an inert state until activated by binding to cell surface glycosaminoglycans (GAGs). AT and HCII thus are allosterically activated toward factor Xa and thrombin, respectively, the final two proteases in the blood coagulation cascade.
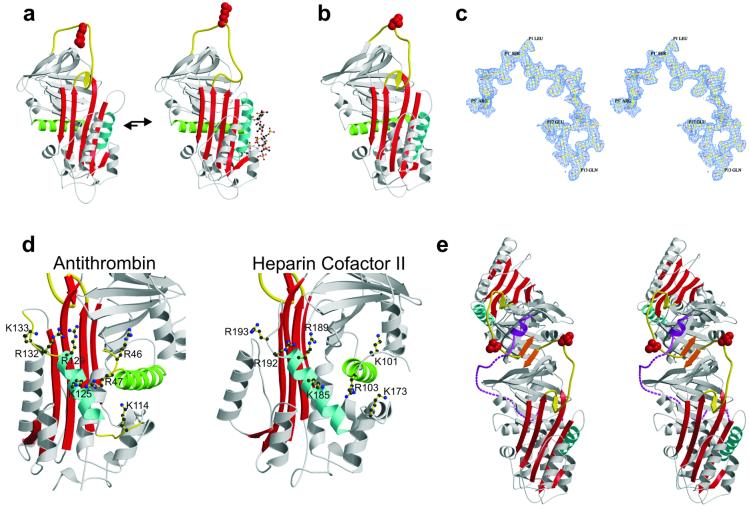
The structure and mechanism of activation of AT has been well characterized. Its fold is similar to that of the rest of the serpin family with the single and important exception that the reactive center loop is constrained through partial incorporation into β-sheet A (refs. 6 and 7; Fig. 1a). The native conformation of AT thus renders it incapable of forming a productive recognition complex (Michaelis complex) with factor Xa, and only as the result the conformational changes brought about through the interaction with a specific heparin pentasaccharide sequence does AT become an efficient inhibitor of factor Xa. Pentasaccharide binding results in secondary structural changes in the heparin binding region and the expulsion of the reactive center loop (8) such that activated AT resembles all the other known native inhibitory serpins with a fully exposed reactive center loop and a five-stranded β-sheet A (Fig. 1a).
Fig 1.
The AT-like structure of native HCII. (a) The native structure of AT is different from all other know serpins with the partial incorporation of the reactive center loop (yellow) into β-sheet A (red), resulting in an unfavorable orientation of the reactive center arginine side chain (red, space-filling). Binding of heparin (ball-and-stick) to helix D (cyan) results in the expulsion of the reactive center loop from β-sheet A and its freeing for the formation of a productive Michaelis complex. (b) The structure of native HCII resembles that of native AT, with its reactive center loop partially inserted into β-sheet A. (c) Stereo depiction of the electron density for the entire reactive center loop contoured at 1σ. (d) The high degree of conservation of heparin binding residues on helix D (cyan) and helix A (green) with AT indicates a similar mode of binding and suggests similar conformational changes in response to binding. (e) A stereo representation of the crystallographic dimer of HCII formed by the antiparallel β-sheet interaction between strands 1C (orange) of each monomer. This contact is the probable cause of the displacement of the acidic N-terminal tail (magenta). The bottom monomer is molecule A.
The reactive center-loop sequence of HCII implies specificity for chymotrypsin, with a leucine in the critical P1 position (where the scissile bond is denoted P1–P1′; ref. 9), yet it is a specific inhibitor of thrombin, which normally requires a P1 arginine. The specificity of HCII for thrombin is conferred by a highly acidic hirudin-like N-terminal tail, which becomes available after GAG binding for interaction with the anion-binding exosite I of thrombin (10–14). A mechanism for HCII activation has been proposed (11, 15) whereby the N-terminal tail is sequestered in the native structure through an interaction with the highly basic heparin binding region, and activation is brought about through the competitive displacement of the acidic tail of HCII by heparin or similar anionic GAGs. Here we describe, in turn, the crystallographic structures of native and S195A thrombin-complexed HCII. The native structure displays unexpected homology to AT, which provides a previously uncharacterized mechanism for sequestration and release of the acidic tail, and the Michaelis complex between HCII and thrombin defines the molecular basis of activation. Together, these structures reveal a multistep mechanism that depends on the sequential contraction and expansion of β-sheet A and demonstrate the degree to which the complicated serpin mechanism of protease inhibition has been exploited by evolution to effect regulatory control.
Methods
Native HCII.
HCII was purified as described (16). Crystals were obtained from Hampton Crystal Screen Cryo 6 (Hampton Research, Riverside, CA) and were generally very thin plates that diffracted poorly. Crystals were shown to contain intact HCII by SDS/PAGE, although some degradation, presumably N-terminal cleavage, was observed in older crystals. A data set was collected on a single frozen crystal at the SRS Daresbury Station 14.1 (Cheshire, U.K.) on an ADSC image plate. The crystal diffraction was nonisotropic, and significant decay was observed during data collection. The data were processed using MOSFLM, SCALA, and TRUNCATE (17) and were of reasonable quality to a resolution limit of 2.35 Å (Table 1). The structure of native HCII was solved by molecular replacement (MolRep; ref. 18) using native plasminogen activator inhibitor-1 (PDB ID code ), native AT (PDB ID code ), latent AT (PDB ID code ), and pentasaccharide-activated AT (PDB ID code ) as search models. Comparison of the relative scores for the molecular replacement solutions clearly indicated that the best solution was for native AT and that two monomers occupied the asymmetric unit. A homology model of HCII was generated on the native AT template by using SWISSMODEL (19), which then was subjected to rigid-body refinement and coordinate refinement with simulated annealing by using CNS (20). The Rfree set was chosen randomly from a subset of reflections unrelated by noncrystallographic symmetry. Iterative rounds of rebuilding in XTALVIEW (21) and refinement with CNS resulted in an Rfactor and Rfree of 20.8 and 24.7, respectively (Table 1). Because the twofold noncrystallographic symmetry axis was nearly crystallographic, averaged, solvent-flattened maps were calculated with 5,284 unobserved reflections regenerated using the program DM (22).
Table 1.
Crystals, data processing, refinement, and models
| Crystals | Native HCII | HCII–S195A-thrombin complex |
| Space group | P21 | P61 |
| Cell dimensions, Å | a = 74.7, b = 80.0, c = 92.2, β = 102.0° | a, b = 152.3, c = 126.81 |
| Solvent content, % | 46 | 72 |
| Data-processing statistics | ||
| Wavelength, Å | 1.488 | 1.488 |
| Resolution, Å | 25.1–2.35 | 29.4–2.2 |
| Total reflections | 112,978 | 475,985 |
| Unique reflections | 39,178 | 77,117 |
| 〈I/σ(I)〉 | 4.0 | 6.5 |
| 〈I〉/σ(〈I〉) | 11.7 | 14.8 |
| Completeness, % | 88.0 | 91.1 |
| Multiplicity | 2.9 | 6.2 |
| Rmerge | 0.14 | 0.08 |
| Model | ||
| No. of protein/water atoms | 6,376/297 | 5,933/208 |
| Average B factor, Å2 | 37.7 | 55.0 |
| Refinement statistics | ||
| Reflections in working/free set | 38,162/994 | 76,094/983 |
| Rfactor/Rfree | 20.8/24.7 | 20.5/21.1 |
| rms deviation of bonds, Å/angles, °, from ideality | 0.006/1.3 | 0.006/1.4 |
| Ramachandran plot; residues in | ||
| Most favored region, % | 90.8 | 86.5 |
| Additionally allowed region, % | 9.1 | 12.1 |
| Generously allowed region, % | 0 | 1.1 |
| Disallowed region, % | 0.1 | 0.3 |
HCII–S195A Thrombin Complex.
S195A thrombin was produced as described (23). The 1:1 complex of S195A thrombin and HCII, concentrated to 8–10 mg/ml, was crystallized in 0.06M NH4Cl and 6% polyethylene glycol 3350. The crystals were large and hexagonal (400 × 400 × 300 μm) but diffracted poorly and decayed rapidly when exposed to X-rays at room temperature. Improved diffraction was achieved through systematic dehydration by the sequential rapid stepping-up of 2-methyl-2,4-pentanediol (MPD) concentration from 0 to 20% in 2% increments. The crystal was flash frozen and data were collected and processed as above. The structure of the complex was solved by molecular replacement using the program MOLREP with our structure of S195A thrombin (PDB ID code ) and with the N-terminal half of native HCII (residues 100–265) as search models. The C-terminal half of HCII (residues 282–480) was subsequently positioned with the solutions for thrombin and the N-terminal fraction fixed. Molecular replacement attempts with native HCII were unsuccessful even with the thrombin solution fixed using both MOLREP and AMORE (24). The final refined model was obtained as described above. Solvent-flattened maps were calculated by using DM because of the unusually high solvent content (72%). Processing and refinement statistics are given in Table 1. Figures were made by using MOLSCRIPT (25), BOBSCRIPT (26), RASTER3D (27), and GRASP (28).
Results and Discussion
Overall Structure of Native HCII.
The overall structure of native HCII is nearly identical to that of native AT, with a six-stranded β-sheet A due to the partial incorporation of the reactive center loop (Fig. 1). Superposition of HCII and AT results in an rms deviation of 1.8 Å for 352 equivalent Cα atoms despite the rather low sequence identity of 26.8%. It is likely that AT and HCII have coincidentally adopted the same native conformation for the same purpose, that of sequestering the reactive center loop so as to render the circulating form nonreactive toward target and nontarget proteases, and that GAG binding will result in expulsion of the reactive center loop of HCII, as with AT (Fig. 1a). One of the major surprises in finding native HCII in this conformation is that the P1 leucine would already render a normal loop-expelled, five-stranded native form inactive toward thrombin. This fact has been demonstrated by studies that show that reactivity toward thrombin requires an intact N terminus even in the presence of GAG (10). In AT, expulsion of the reactive center loop after heparin binding also has the effect of increasing the affinity for heparin by 1,000-fold and globally altering its surface properties. The AT-like native conformation of HCII thus provides the basis for a hypothesis for the mechanism of its GAG activation, where the acidic N terminus is released because of the global conformational changes associated with GAG-induced expulsion of the partially inserted reactive center loop.
The Heparin Binding Site.
The heparin-binding residues of HCII were identified initially by sequence alignment with AT and have since been confirmed through the study of variants to be Lys-173, Arg-184, Lys-185, Arg-189, Arg-192, and Arg-193 on or near helix D (15). Fig. 1d compares the heparin binding sites of AT and HCII, with key residues indicated. The critical residues involved in the tight binding of the heparin pentasaccharide by AT have been demonstrated by both structural and biochemical studies to include Lys-114, Lys-125, and Arg-129, whereas Arg-132, Lys-133, and Lys-136 contribute to the binding of longer heparin chains (8, 29–33). The conservation in HCII of the key residues involved in the interaction of the pentasaccharide with AT supports a similar mechanism of GAG binding by HCII. Further evidence for a common binding mechanism comes from the conservation of other AT residues known to interact with heparin such as Arg-46 and Arg-47 on helix A (34), which correspond to Lys-101 and Arg-103 in HCII. The structure of the heparin binding region of native HCII displays an inherent flexibility in loops containing basic residues critical for GAG binding and suggests a structural response to GAG binding similar to that observed for AT.
The Acidic Tail.
Although allostery is implied by analogy to AT, the position for the majority of the acidic tail is undefined in the structure of native HCII. Of the ≈160 amino acids in the N-terminal tails of the two molecules comprising the crystallographic asymmetric unit (Fig. 1e), only 11 can be unequivocally placed in density (Leu-61–Asp-71). It was not possible to assign the observed portion of the acidic tail to a particular molecule in the asymmetric unit based on connectivity of electron density, but it most likely belongs to molecule A because of limitations of available threading pathways through the crystal.
The fact that the acidic tail is not ordered in the crystallographic structure of HCII indicates that it is flexible within the crystal. It is unlikely that the acidic tail is flexible in solution, because it is not proteolytically susceptible in the native state (35). Thus, the native position of the acidic tail has been perturbed by crystal contacts. The proposal that the tail interacts with the heparin binding site located on helix D is not supported by this structure, because there are no crystal contacts on helix D, and no unassigned electron density is found in this region. The most stable interaction within the crystal is that between the two molecules that makeup the asymmetric unit (Fig. 1e). This antiparallel β-sheet interaction between strands 1 of sheet C (orange) results in a continuous sheet C from molecule A to molecule B. The stability of such an interface has been demonstrated by the observation of similar interactions in crystals and in solution for AT (7, 36) and plasminogen activator inhibitor-1 (37, 38). The interface between the two monomers also corresponds to the position of the fragment of the acidic tail observed in the structure. It thus is likely that the dimerization of monomers to form the asymmetric unit is required for crystallization and results in an unfavorable apposition of the two acidic tails, which would place a portion of the acidic tail in native HCII in contact with sheet C.
Overall Structure of the HCII–S195A Thrombin Michaelis Complex.
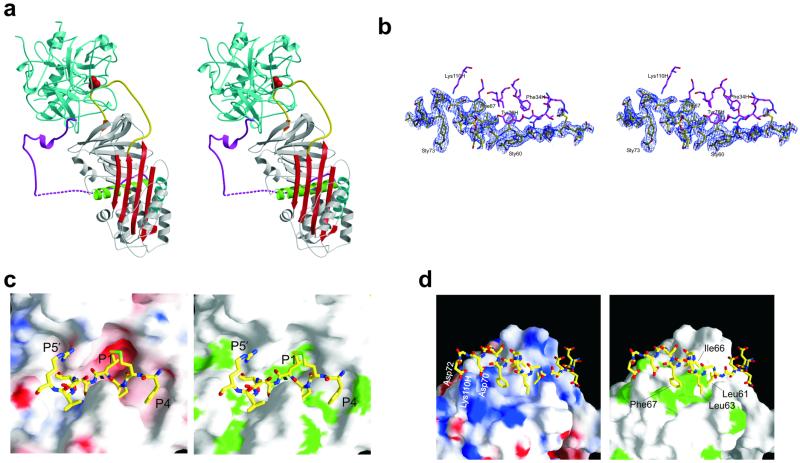
The prediction from the native structure alone is that the activated form of HCII will have an expelled reactive center loop and thus a five-stranded β-sheet A, as occurs in AT activation (Fig. 1a), which was verified by the solution of the crystal structure of HCII in complex with the enzymatically inert S195A variant of human thrombin (Fig. 2a). Thrombin is oriented at the top of the serpin, in the classic view, and slightly behind and to the left with the heparin binding site of thrombin (exosite II) aligned with the helix D of HCII to allow heparin bridging. Although this complex represents a Michaelis complex between a serpin and a protease, it cannot be generalized to all serpin–protease Michaelis complexes because of the intimacy of the interface. NMR studies on the antitrypsin–anhydrotrypsin complex (39) and a recently published structure of the Manduca Sexta serpin–S195A trypsin complex (40) demonstrate a flexible Michaelis complex where the serpin and the protease rotate as independent bodies. In these cases, the specificity is determined wholly by the reactive center loop sequence and in particular the P1 residue. Thus, to render the Michaelis complexes between antitrypsin or the Manduca Sexta serpin and trypsin stable enough for NMR and crystallography, the P1 residue was changed from Met → Arg and from Leu → Lys, respectively. The ability of HCII to form a productive Michaelis complex with thrombin depends on exosite interactions and thus can accommodate a leucine at the P1 position. In the HCII–S195A thrombin complex, thrombin is effectively docked on the plane of sheet C and makes simultaneous contacts with the acidic tail and the reactive center loop, resulting in a complex that, in solution, would rotate as a rigid body.
Fig 2.
Crystallographic structure of the HCII–thrombin Michaelis complex. (a) A stereo representation of the Michaelis complex between S195A thrombin (cyan) and HCII (colored as described for Fig. 1), with the γ-loop in front and the 60-insertion loop behind the reactive center loop. (b) Stereo representation of the electron density covering the portion of the acidic tail (yellow) that interacts with thrombin (magenta), contoured at 1σ. (c) The subsite interactions between the reactive center residues of HCII (rods) and the active site cleft of thrombin (surface representation) are extensive and complementary in both electrostatic (Left, negative potential is red, and positive is blue) and hydrophobic (Right, green for hydrophobic side chains) properties. (d) The interaction between exosite I of thrombin (surface representation as described for c) and the hirudin-like N-terminal tail of HCII (rods, the side chains of residues not interacting with thrombin were removed for clarity) is primarily hydrophobic. The only ionic interactions are between Asp-70 and Asp-72 of HCII and L110H of thrombin. Sulfated tyrosines, 60 and 73, make no contacts with thrombin and are not shown.
It is clear from this structure why the reactive center loop needs to be released from the constraints of partial β-sheet incorporation to form the Michaelis complex with thrombin; in striking contrast to the structure of the Manduca Sexta serpin–trypsin Michaelis complex (40), the reactive center loop has been stretched to the maximal extent to simultaneously make subsite contacts with thrombin within the reactive center loop and elsewhere in the molecule. The reactive center loop forms a tight turn in the active site cleft of thrombin, and the hirudin domain, sandwiched between HCII and thrombin, participates in extensive hydrophobic and two electrostatic interactions with exosite I of thrombin. The position of the hirudin tail is well defined in the structure of the complex, with uninterrupted density from residue 54 to 83 (Fig. 2b) and from 94 to the N terminus of helix A. The tail interacts with thrombin from residue 56 to 72 and then is entirely free from noncrystallographic interactions until residue 94. The striking flexibility of the intervening portion of the tail is evident in the stereo representation of the complex (Fig. 2a) and is in keeping with the degree of mobility required to allow the extraordinary displacement of thrombin that is central to the serpin inhibitory mechanism.
The Interface.
The Michaelis complex between HCII and thrombin differs from previously characterized serpin–protease Michaelis complexes in the extent of subsite interactions outside the reactive center loop. It is the strength of these exosite interactions that accounts for the high rate of thrombin inhibition by GAG-activated HCII. The interaction interface can be divided into three regions of HCII that make important contacts with thrombin: (i) the reactive center loop, (ii) sheet C and elsewhere, and (iii) the acidic tail.
The Reactive Center Loop.
The reactive center loop of native HCII is constrained by its partial incorporation into β-sheet A, which was a surprise finding that is explained nicely by the structure of HCII–S195A thrombin complex; in order for thrombin to simultaneously bind to the reactive center and form interactions elsewhere, the reactive center loop must be expelled and indeed stretched to the fullest extent possible. Thus, loop expulsion after GAG binding is a necessary part of the activation mechanism. The interactions with thrombin involve residues P4–P5′ (Phe-441–Arg-449), which form a tight loop within the active site. The interactions are illustrated by surface representations of the active site of thrombin colored according to electrostatic and hydrophobic properties in Fig. 2c.
Sheet C and Elsewhere.
Thrombin differs most significantly from other serine proteases in the extensions at residues 60 and 147, known as the 60-insertion and γ-loops (41), which restrict access of substrates to the active site. The 60-insertion loop makes several contacts within the reactive center loop, but it also makes contacts elsewhere on the body of HCII. A critical contact is the salt bridge between Asp60E of thrombin and Arg-369 of HCII. The γ-loop makes no favorable contacts within the reactive center loop of HCII but has extensive interactions with residues on or adjacent to β-sheet C. Trp147A is buried in a hydrophobic pocket formed by Met-288, Gln-308, His-290 of HCII, and the backbone of the γ-loop itself. Asn147D forms hydrogen bonds with main chain atoms of Glu-287, Thr-289, and Val-286 of HCII. These interactions, although not expected to contribute greatly to the binding energy for the HCII–thrombin Michaelis complex, illustrate the strategy of HCII in accommodating these bulky thrombin loops, which is in contrast to AT, which simply possesses a longer reactive center loop.
The Acidic Tail.
The acidic tail of HCII contains two so-called hirudin domains, 56EDDDY*LD62 and 69EDDDY*ID75, each of which contains several acidic amino acids and a sulfated tyrosine. The interface between the acidic tail and thrombin in the structure of the HCII–S195A thrombin complex is striking in that there are only two ionic interactions (Asp-70 and Asp-72 with Leu110H). Much like the hirudin–thrombin complex (PDB ID code ; ref. 42), the majority of the contacts are hydrophobic (Fig. 2d). The anion-binding exosite I contains a very hydrophobic face made up of Phe-34, Leu-65, Arg-67, Tyr-76, Ile-82, and Met-84. In the HCII–thrombin Michaelis complex, this hydrophobic patch is well covered by an amphipathic helix, which lies between the two highly acidic hirudin domains, and includes residues Leu-61, Leu-63, Ile-66, and Phe-67. The analogous region on hirudin does not form a helix but contains a similar complement of hydrophobic residues. The N- to C-terminal orientation of the acidic tail on exosite I also is the same as for hirudin.
The N-terminal hirudin domain is effectively sandwiched in between HCII and thrombin, and its only ionic interaction is intramolecular (Asp-58–Arg-295). The sulfated tyrosine (Sty60) is also making intramolecular contacts with the main chain amide hydrogen of Met-344, which is the only contact made for either of the two sulfated tyrosines in HCII. The sulfated tyrosines thus may be more important for intramolecular interactions that sequester the acidic tail in the native state than for the stabilization of the HCII–thrombin complex. All other interactions are main chain or side chain hydrogen bonds involving Glu-56, Asp-57, Asp-59, and Leu-61.
Allosteric Activation of HCII.
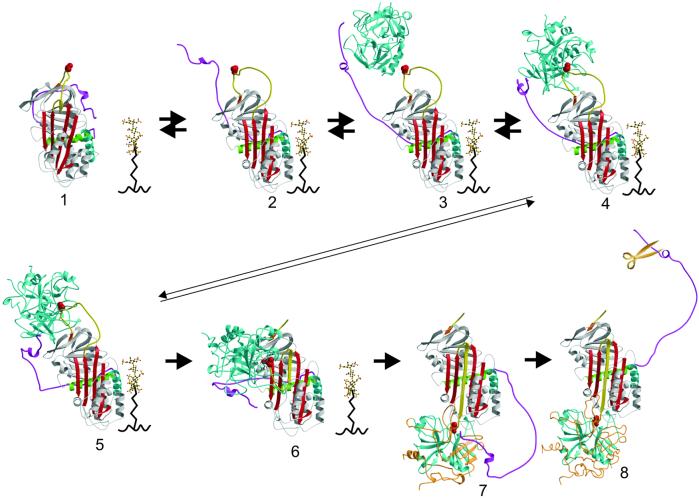
How the acidic tail is freed by heparin-like GAGs for the formation of the initial complex with thrombin depends on where the tail is in native HCII. Although the native structure does not provide an unequivocal position for the tail, it does provide clues as to where it may reside. First, it is not where it appears in the structure of the native HCII dimer (Fig. 1e), because the observed portion of the tail interacts only weakly with the body of HCII. Second, it is not bound to the GAG-binding region (helix D) as predicted (10), because this part of the molecule is free of crystal contacts in the native crystal, and no electron density is observed. Third, the identical orientation of the N-terminal loop of helix A (residues 94–101) implies an initial position very similar to that found in the structure of the HCII–S195A thrombin complex. Thus, a model for the position of the tail in native HCII was created based on the above considerations and the assumption that s1C dimerization disrupts the position of the tail in the crystal of native HCII. Additionally, a recent study has demonstrated a re-formable disulfide bond for the P52C, F195C HCII variant,∥ which imposes a strict distance constraint. A good model for the position of the acidic tail in native HCII would require both charge complementarity and the burying of the hydrophobic face of the amphipathic helix against a hydrophobic patch on the body of HCII. These factors are nicely but not uniquely satisfied by the model shown in Fig. 3 (model 1). This model suggests that the acidic tail in native HCII would interfere with thrombin docking and is consistent with the higher basal rate of thrombin inhibition for the tail-deleted HCII variant (43). An attractive alternative to the positioning of the tail illustrated in the Fig. 3 would be to place it in contact with the reactive center loop itself. Such a position would explain why the acidic tail is susceptible to proteolysis after cleavage of the reactive center loop (44). In either case, the extended nature of the predicted interactions between the acidic tail and the body of HCII is consistent with an allosteric mechanism for its release. The conformational changes associated with the expulsion of the reactive center loop of HCII are manifested in gross changes in surface shape and properties of HCII. How these conformational changes could release the acidic tail is illustrated by the comparison of the surface electrostatic and hydrophobic contours of the region adjacent to the GAG-binding site and the thrombin-docking region. A video depiction that flips between the two conformations illustrates the magnitude of these changes in shape and properties (Movie 1, which is published as supporting information on the PNAS web site, www.pnas.org).
Fig 3.
The sequential mechanism of GAG-mediated thrombin inhibition by HCII (as described in Results and Discussion). For a video of this mechanism, see Movie 2.
The HCII Mechanism.
The structures presented here, combined with those of AT and the final serpin–protease complex, reveal a multistep allosteric mechanism for HCII (Fig. 3). A video, based on these frames, is shown in Movie 2, which is published as supporting information on the PNAS web site. Briefly, the binding of HCII to GAG at the cell surface initiates conformational changes including the expulsion of the reactive center loop, closure of β-sheet A, and the consequent release of the acidic tail (Fig. 3, models 1 and 2). Through an induced-fit mechanism analogous to that of AT, the five-stranded, activated conformation of HCII binds its specific GAG with a greater affinity. The hirudin-like tail of HCII is now free to recruit and tether thrombin (model 3). Similarly, the reactive center loop is free to make subsite contacts within the active site cleft of thrombin (model 4), followed by a tilting movement that allows the thrombin and the bound hirudin domain to form intimate contacts with the body of HCII (model 5). The consequent stability of the Michaelis complex allows for progression into the proteolytic cycle. At the acyl-enzyme intermediate step, HCII rapidly incorporates its reactive center loop into β-sheet A, and the tethered thrombin, still bound to the hirudin-like tail, is translocated to the opposite pole of HCII (models 6 and 7). Steric clashes induced by the formation of the final complex disrupt the entirety of exosite I (45, 46), effecting the re-release of the acidic tail (model 8), and conformational changes associated with full incorporation of the reactive center loop prevent the re-binding of the acidic tail to the body of HCII. The freed tail then can be cleaved (35) by resident proteases releasing the peptide corresponding to residues 49–60 to stimulate leukocyte chemotaxis (47–49). Finally, expansion of β-sheet A results in a reduction in affinity for GAG (44), and thus the HCII–thrombin complex is released from the cell surface for catabolic destruction (50, 51), enabling the GAG to be recycled for further catalysis.
The structures presented here illustrate the extent of regulatory control afforded by the serpin inhibitory mechanism and go a long way in explaining why serpins control the critical proteolytic pathways in the higher organisms. The conformational changes associated with the expansion and contraction of the central β-sheet A define all aspects of HCII activity from cofactor binding and thrombin recognition to protease inhibition and cell signaling.
Supplementary Material
Acknowledgments
We are grateful to Randy Read and Navraj Pannu for assistance throughout. Funding was provided by the National Institutes of Health (to F.C.C., C.T.E., and J.A.H.), the Wellcome Trust (to R.W.C.), and the Medical Research Council (to J.A.H.). T.P.B. was supported by the PPP foundation.
Abbreviations
AT, antithrombin
HCII, heparin cofactor II
GAG, glycosaminoglycan
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes and for native and S195A thrombin-complexed HCII, respectively).
Brinkmeyer, S., Eckert, R. & Ragg, H. (2001) Thromb. Haemostasis Suppl. (abstr.).
References
- 1.Gettins P. G. W., Patston, P. A. & Olson, S. T., (1996) Serpins: Structure, Function, and Biology (Landes, Austin, TX).
- 2.Huntington J. A. & Carrell, R. W. (1901) Sci. Prog. 84, 125-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huntington J. A., Read, R. J. & Carrell, R. W. (2000) Nature (London) 407, 923-926. [DOI] [PubMed] [Google Scholar]
- 4.Atchley W. R., Lokot, T., Wollenberg, K., Dress, A. & Ragg, H. (2001) Mol. Biol. Evol. 18, 1502-1511. [DOI] [PubMed] [Google Scholar]
- 5.Ragg H., Lokot, T., Kamp, P. B., Atchley, W. R. & Dress, A. (2001) Mol. Biol. Evol. 18, 577-584. [DOI] [PubMed] [Google Scholar]
- 6.Schreuder H. A., de Boer, B., Dijkema, R., Mulders, J., Theunissen, H. J., Grootenhuis, P. D. & Hol, W. G. (1994) Nat. Struct. Biol. 1, 48-54. [DOI] [PubMed] [Google Scholar]
- 7.Carrell R. W., Stein, P. E., Fermi, G. & Wardell, M. R. (1994) Structure (London) 2, 257-270. [DOI] [PubMed] [Google Scholar]
- 8.Jin L., Abrahams, J. P., Skinner, R., Petitou, M., Pike, R. N. & Carrell, R. W. (1997) Proc. Natl. Acad. Sci. USA 94, 14683-14688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schechter I. & Berger, A. (1967) Biochem. Biophys. Res. Commun. 27, 157-162. [DOI] [PubMed] [Google Scholar]
- 10.Van Deerlin V. M. & Tollefsen, D. M. (1991) J. Biol. Chem. 266, 20223-20231. [PubMed] [Google Scholar]
- 11.Ragg H., Ulshofer, T. & Gerewitz, J. (1990) J. Biol. Chem. 265, 5211-5218. [PubMed] [Google Scholar]
- 12.Myles T., Church, F. C., Whinna, H. C., Monard, D. & Stone, S. R. (1998) J. Biol. Chem. 273, 31203-31208. [DOI] [PubMed] [Google Scholar]
- 13.Rogers S. J., Pratt, C. W., Whinna, H. C. & Church, F. C. (1992) J. Biol. Chem. 267, 3613-3617. [PubMed] [Google Scholar]
- 14.Sheehan J. P., Wu, Q., Tollefsen, D. M. & Sadler, J. E. (1993) J. Biol. Chem. 268, 3639-3645. [PubMed] [Google Scholar]
- 15.Tollefsen D. M. (1997) Adv. Exp. Med. Biol. 425, 35-44. [DOI] [PubMed] [Google Scholar]
- 16.Griffith M. J., Noyes, C. M. & Church, F. C. (1985) J. Biol. Chem. 260, 2218-2225. [PubMed] [Google Scholar]
- 17.Leslie A. W. G., (1992) Joint CCP4 and ESF-EACMB Newsletter on Protein Crystallography (Daresbury Laboratory, Warrington, U.K.), pp. 26.
- 18.Vagin A. & Teplyakov, A. (2000) Acta Crystallogr. D Biol. Crystallogr. 56, 1622-1624. [DOI] [PubMed] [Google Scholar]
- 19.Guex N. & Peitsch, M. C. (1997) Electrophoresis 18, 2714-2723. [DOI] [PubMed] [Google Scholar]
- 20.Brunger A. T., Adams, P. D., Clore, G. M., Delano, W. L., Gros, P., GrosseKunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 21.McRee D. E. (1992) J. Mol. Graph. 10, 44-46. [Google Scholar]
- 22.Cowtan K. & Main, P. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 487-493. [DOI] [PubMed] [Google Scholar]
- 23.Ye J., Rezaie, A. R. & Esmon, C. T. (1994) J. Biol. Chem. 269, 17965-17970. [PubMed] [Google Scholar]
- 24.Navaza J. (1994) Acta Crystallogr. A 50, 157-163. [Google Scholar]
- 25.Kraulis P. J. (1991) J. Appl. Crystallogr. 24, 946-950. [Google Scholar]
- 26.Esnouf R. M. (1997) J. Mol. Graph. Modell. 15, 132. [DOI] [PubMed] [Google Scholar]
- 27.Merritt E. A. & Murphy, M. E. P. (1994) Acta Crystallogr. D 50, 869-873. [DOI] [PubMed] [Google Scholar]
- 28.Nicholls A. & Honig, B. (1991) J. Comput. Chem. 12, 435-445. [Google Scholar]
- 29.Arocas V., Bock, S. C., Raja, S., Olson, S. T. & Bjork, I. (2001) J. Biol. Chem. 276, 43809-43817. [DOI] [PubMed] [Google Scholar]
- 30.Schnedin-Weiss S., Desai, U. R., Bock, S. C., Gettins, P. G. W., Olson, S. T. & Bjork, I. (2002) Biochemistry 41, 4779-4788. [DOI] [PubMed] [Google Scholar]
- 31.Desai U., Swanson, R., Bock, S. C., Bjork, I. & Olson, S. T. (2000) J. Biol. Chem. 275, 18976-18984. [DOI] [PubMed] [Google Scholar]
- 32.Meagher J. L., Huntington, J. A., Fan, B. & Gettins, P. G. (1996) J. Biol. Chem. 271, 29353-29358. [DOI] [PubMed] [Google Scholar]
- 33.Arocas V., Turk, B., Bock, S. C., Olson, S. T. & Bjork, I. (2000) Biochemistry 39, 8512-8518. [DOI] [PubMed] [Google Scholar]
- 34.Arocas V., Bock, S. C., Olson, S. T. & Bjork, I. (1999) Biochemistry 38, 10196-10204. [DOI] [PubMed] [Google Scholar]
- 35.Pratt C. W., Tobin, R. B. & Church, F. C. (1990) J. Biol. Chem. 265, 6092-6097. [PubMed] [Google Scholar]
- 36.Zhou A., Huntington, J. A. & Carrell, R. W. (1999) Blood 94, 3388-3396. [PubMed] [Google Scholar]
- 37.Sharp A. M., Stein, P. E., Pannu, N. S., Carrell, R. W., Berkenpas, M. B., Ginsburg, D., Lawrence, D. A. & Read, R. J. (1999) Structure Fold. Des. 7, 111-118. [DOI] [PubMed] [Google Scholar]
- 38.Zhou A., Faint, R., Charlton, P., Dafforn, T. R., Carrell, R. W. & Lomas, D. A. (2001) J. Biol. Chem. 276, 9115-9122. [DOI] [PubMed] [Google Scholar]
- 39.Peterson F. C., Gordon, N. C. & Gettins, P. G. (2000) Biochemistry 39, 11884-11892. [DOI] [PubMed] [Google Scholar]
- 40.Ye S., Chech, A. L., Belmares, R., Bergstrom, R. C., Tong, Y., Corey, D. R., Kanost, M. R. & Goldsmith, E. J. (2001) Nat. Struct. Biol. 8, 979-983. [DOI] [PubMed] [Google Scholar]
- 41.Stubbs M. T. & Bode, W. (1993) Thromb. Res. 69, 1-58. [DOI] [PubMed] [Google Scholar]
- 42.Stubbs M. T., Oschkinat, H., Mayr, I., Huber, R., Angliker, H., Stone, S. R. & Bode, W. (1992) Eur. J. Biochem. 206, 187-195. [DOI] [PubMed] [Google Scholar]
- 43.Liaw P. C., Austin, R. C., Fredenburgh, J. C., Stafford, A. R. & Weitz, J. I. (1999) J. Biol. Chem. 274, 27597-27604. [DOI] [PubMed] [Google Scholar]
- 44.Maekawa H., Sato, H. & Tollefsen, D. M. (2000) Thromb. Res. 100, 443-451. [DOI] [PubMed] [Google Scholar]
- 45.Bock P. E., Olson, S. T. & Bjork, I. (1997) J. Biol. Chem. 272, 19837-19845. [DOI] [PubMed] [Google Scholar]
- 46.Fredenburgh J. C., Stafford, A. R. & Weitz, J. I. (2001) J. Biol. Chem. 276, 44828-44834. [DOI] [PubMed] [Google Scholar]
- 47.Church F. C., Pratt, C. W. & Hoffman, M. (1991) J. Biol. Chem. 266, 704-709. [PubMed] [Google Scholar]
- 48.Hoffman M., Pratt, C. W., Brown, R. L. & Church, F. C. (1989) Blood 73, 1682-1685. [PubMed] [Google Scholar]
- 49.Hoffman M., Pratt, C. W., Corbin, L. W. & Church, F. C. (1990) J. Leukocyte Biol. 48, 156-162. [DOI] [PubMed] [Google Scholar]
- 50.Maekawa H. & Tollefsen, D. M. (1996) J. Biol. Chem. 271, 18604-18609. [DOI] [PubMed] [Google Scholar]
- 51.Kounnas M. Z., Church, F. C., Argraves, W. S. & Strickland, D. K. (1996) J. Biol. Chem. 271, 6523-6529. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.