Abstract
Ca2+, pheromones, sweet taste compounds, and the main neurotransmitters glutamate and γ-aminobutyric acid activate G protein-coupled receptors (GPCRs) that constitute the GPCR family 3. These receptors are dimers, and each subunit has a large extracellular domain called a Venus flytrap module (VFTM), where agonists bind. This module is connected to a heptahelical domain that activates G proteins. Recently, the structure of the dimer of mGlu1 VFTMs revealed two important conformational changes resulting from glutamate binding. First, agonists can stabilize a closed state of at least one VFTM in the dimer. Second, the relative orientation of the two VFTMs in the dimer is different in the presence of glutamate, such that their C-terminal ends (which are connected to the G protein-activating heptahelical domain) become closer by more than 20 Å. This latter change in orientation has been proposed to play a key role in receptor activation. To elucidate the respective role of VFTM closure and the change in orientation of the VFTMs in family 3 GPCR activation, we analyzed the mechanism of action of the mGlu8 receptor antagonists ACPT-II and MAP4. Molecular modeling studies suggest that these two compounds prevent the closure of the mGlu8 VFTM because of ionic and steric hindrance, respectively. We show here that the replacement of the residues responsible for these hindrances (Asp-309 and Tyr-227, respectively) by Ala allows ACPT-II or MAP4 to fully activate the receptors. These data are consistent with the requirement of the VFTM closure for family 3 GPCR activation.
G protein-coupled receptors (GPCRs) represent the major gene family in mammalian genomes. They are involved in the action of many types of extracellular signals from photon to large proteins, from external sensory molecules to hormones and neurotransmitters (1). Among the various families of GPCRs, family 3 receptors are activated by important compounds such as Ca2+, pheromones, sweet molecules, and the main neurotransmitters γ-aminobutyric acid and glutamate. As other GPCRs, family 3 receptors have a heptahelical domain (HD) responsible for G protein activation (2). However, they possess a large, extracellular domain structurally similar to bacterial periplasmic-binding proteins that contain the agonist-binding site (3–10). As clearly shown by the solved x-ray structure of the glutamate-binding domain of the metabotropic glutamate receptor type 1 (mGlu1 receptor) (11), this domain is constituted of two lobes separated by a large cleft on which agonists bind and is called a Venus flytrap module (VFTM). A second feature of family 3 receptors is that they all form dimers, either homodimers (12–15) or heterodimers (16–18).
How does the binding of agonists in the extracellular VFTMs lead to the activation of the HD? Important new information has been obtained as a result of the determination of the crystal structure of the dimer of VFTMs of the mGlu1 receptor with and without bound glutamate or the mGlu1 antagonist α-methyl-4-carboxyphenylglycine (MCPG) (11, 19). These studies revealed two major conformational changes resulting from agonist binding. A first one is the closure of at least one VFTM in the dimer, as expected from modeling studies of other family 3 GPCRs (6, 8–10, 20). Indeed, glutamate binds to lobe I within the cleft that separates both lobes and also can interact with residues from lobe II leading to the stabilization of a closed state. The second major change in conformation is the rotation of one VFTM relative to the other, such that the C-terminal ends of each VFTM in the dimer become closer by more than 20 Å (11). This may lead to a different interaction of the HDs within the dimer, possibly stabilizing their active conformation. Such a possibility fits nicely with recent data obtained with the γ-aminobutyric acid type B heteromeric receptor (21, 22). Moreover, a bringing together of the C-terminal ends of each extracellular domain of the dimeric guanylate cyclase natriuretic peptide receptors also has been proposed to play a pivotal role in receptor activation (23, 24). Of interest, the extracellular domain of these receptors also corresponds to a VFTM.
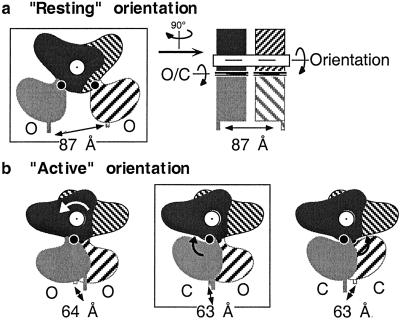
Although experimental data support the importance of the change in the relative orientation of the VFTMs in receptor activation, the possible role of VFTM closure is still unknown. Indeed, analysis of the possible quaternary structure of the dimer of mGlu1 VFTMs shows that the same distance between the C-terminal ends of the two VFTMs can be observed whether each of the modules is in a closed or open conformation as long as the relative orientation of the VFTMs is maintained (Fig. 1). One therefore may wonder whether the closure of one VFTM is required for the change in the relative orientation of the VFTMs. Indeed, in the case of the natriuretic peptide receptors, the agonist induces the bringing together of the C-terminal ends even though it stabilizes both VFTMs in an open conformation (23). It is therefore possible that the closure of the VFTM of family 3 GPCRs serves to control ligand affinity (25), whereas the change in the relative orientation of the two VFTMs in the dimer is the real “motor” for receptor activation.
Fig 1.
Distance between the C-terminal ends of the VFTMs in the dimeric mGlu1 receptor extracellular domain depends on the relative orientation of the VFTMs. (a) Schemes depict the dimeric VFTMs in their “resting” orientation with both VFTMs in an open state, as observed in the crystal structure of the empty form (PDB ID code ) or the MCPG-bound form (PDB ID code ). One VFTM is in the front plane (gray), whereas the other is in the back plane (hatched). Lobe I is darker than lobe II. The right scheme corresponds to a view from the right side of the left scheme and highlights the main axis responsible for the change in conformation of this dimer. The axis for the rotation of one VFTM relative to the other is indicated in white, and the axis in each VFTM responsible for their closure is indicated in black. Models for the dimeric VFTMs in their resting orientation, with one or both modules in the closed state, reveal a similar 87-Å distance between their C-terminal ends (not shown). (b) Schemes represent the different possible conformations of the dimeric VFTMs in their “active” orientation, with both modules in an open state (Left) or one (Center) or both (Right) modules in the closed state. The latter two forms have been observed in crystals with bound glutamate (PDB ID code ) and glutamate and Gd3+ (PDB ID code ), respectively. The distances between the C-terminal ends of each module (α-carbon of Ile-512) in the resolved structures (boxed schemes) or 3D models are indicated. The models were obtained from the superposition of lobe I of the indicated VFTM conformers on lobe I of each subunit of the proposed resting (PDB ID code ) or active (PDB ID code ) dimers.
Understanding the mechanism of action of competitive antagonists could shed some light on this important issue. Here, we show that steric and/or ionic hindrance revealed by three-dimensional (3D) models of antagonists positioned in a closed form of the mGlu8 VFTM can be removed by mutagenesis, making these antagonists able to fully activate the receptor. These data highlight the importance of a closed form of at least one VFTM for the activation of family 3 GPCRs.
Materials and Methods
Materials.
Glutamate acid was purchased from Sigma. (1R,3R,4S)-1-aminocyclopentane-1,3,4-tricarboxylic acid (ACPT-II), (S)-2-amino-4-phosphonobutanoic acid (L-AP4), (S)-2-amino-2-methyl-4-phosphonobutanoic acid (MAP4), (2S,2′R,3′R)-2-(2′,3′-dicarboxycyclopropyl) glycine (DCG-IV), and (αS)-α-amino-α-[(1S,2S)-2-carboxycyclopropyl]-9H-xanthine-9-propanoic acid (LY341495) were purchased from Tocris Neuramin (Bristol, U.K.). Glutamate pyruvate transaminase was from Roche. FBS, culture medium, and other solutions used for cell culture were from GIBCO/BRL. [3H]Myoinositol (23.4 Ci/mol) was purchased from Perkin–Elmer.
Molecular Modeling Studies.
mGlu8-wt (wild type), -D309A and -Y227F, -Y227A VFTM 3D models were built by comparative modeling with MODELER v.5.0 (INSIGHT II 2000; Accelrys, Orsay, France). They were generated by using the coordinates of the mGlu1 VFTM closed form bound with glutamate (PDB ID code :A) and a sequence alignment described (6, 20). Glutamate and L-AP4 bioactive conformations (26) were positioned manually in the mGlu8-wt binding site according to :A and submitted to energy minimization (Steepest Descent convergence 2 kcal/mol/A; Conjugate Gradient convergence 0.1 kcal/mol/A) (20). This was performed by using the DISCOVER 3.00 calculation engine with the CFF force-field (insight ii; Accelrys). The nonbond cut-off method and dielectric constant were set up to cell-multipole and distance dependent (ɛ = 1R), respectively. Putative binding conformations of mGlu8-wt antagonists ACPT-II and MAP4 then were superimposed on the mGlu8-wt agonists. The same ACPT-II and MAP4 conformations also were positioned in mGlu8-D309A and -Y227F, -Y227A mutants, respectively, and were submitted to the protocol of minimization described above.
Construction of mGlu8a Receptor Mutants.
The N-terminal hemagglutinin (HA) epitope-tagged mGlu8a receptor expression plasmid (pRKG8a-NHA) was constructed by replacing the first 34 residues of the mGlu8a sequence being replaced by the mGlu5 signal peptide followed by the HA epitope, as described (27). Single amino acid replacement was carried out on pRKG8a-NHA plasmid by the QuickChange method (Stratagene) according to the manufacturer's instructions. The presence of each mutation of interest and the absence of undesired ones were confirmed by DNA sequencing.
Culture and Transfection of HEK 293 Cells.
HEK 293 cells were cultured in DMEM supplemented with 10% FCS and transfected by electroporation as described, with 14 μg carrier DNA (pRK), plasmid DNA containing mGlu8a or mGlu8a mutants (10 μg), and 10 × 106 cells (28). To enable coupling of mGlu8a and mGlu8a mutant receptors to phospholipase C, the receptors were coexpressed with the chimeric G protein α-subunit Gqi9 (4 μg) (29–31).
Determination of Accumulation of Inositol Phosphates (IPs).
Determination of IP accumulation in transfected cells was performed after labeling the cells overnight with [3H]myoinositol (23.4 Ci/mol) as described (28, 32). Curves were fitted with kaleidagraph software, using the equation y = [(ymax-ymin)/(1 + (x/EC50)nH)] + ymin.
Immunoblotting Analysis.
Twenty hours after transfection, cells were washed with PBS (Ca2+- and Mg2+-free) and harvested. The cell pellet was resuspended in 1 ml of ice-cold lysis buffer [50 mM Tris⋅HCl, pH 7.5/50 mM NaCl/protease inhibitor mixture (Roche)] and homogenized. Cellular debris was discarded by centrifugation at 3,000 rpm for 10 min at 4°C. The supernatant was centrifuged at 44,000 g for 20 min at 4°C, and the membranes, recovered in the pellet fraction, were resuspended in 50 μl of lysis buffer. Protein concentration was measured with Bio-Rad Protein Assay. For each sample, 50 μg of total protein was subjected to SDS/PAGE by using 7.5% polyacrylamide gels, transferred to nitrocellulose membrane (Hybond-C; Amersham Pharmacia), and probed with mAb 12CA5 (Roche) at 0.1 μg/ml. Proteins were visualized by chemiluminescence (West Pico; Pierce).
Immunofluorescence.
Twenty-four hours after transfection, HEK-293 cells plated onto glass coverslips were washed twice with PBS and incubated for 90 min at 37°C with monoclonal mouse 12CA5 at 1.3 μg/ml in PBS/gelatin (0.2%), as described (27). For detection, Cy3 secondary antibody (Jackson ImmunoResearch) was used at 1:1,000. Coverslips were mounted and observed on an upright Axiophot 2 Zeiss microscope.
Results
Choosing mGlu Antagonists to be Studied.
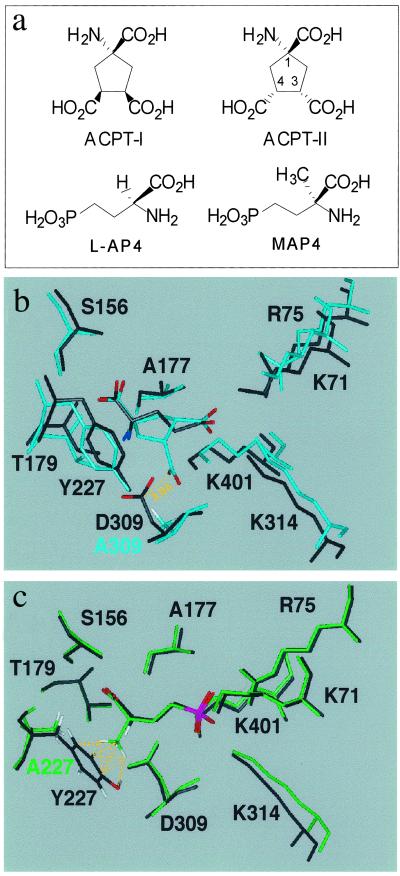
Our actual knowledge of mGlu receptor ligand interaction in the VFTM of the family 3 GPCRs is that the ligand interacts with residues of lobe I and interacts further with residues of lobe II on closure of the VFTM. This is consistent with the observed bound glutamate in the closed and open conformation of the mGlu1 VFTM (PDB ID codes :A and B, respectively) (11). It has been proposed that competitive antagonists also bind to lobe I, like agonists, but prevent the formation of a fully closed state. Indeed, the recent determination of the structure of the mGlu1 VFTM with bound MCPG, an mGlu1 antagonist, revealed that this large compound interacts with both lobes within an open conformation of the VFTM (19). However, many competitive antagonists are derivatives of mGlu receptor agonists in which only small groups have been added. For instance, this is the case with MAP4, the α-methyl derivative of group III agonist L-AP4 (Fig. 2a). Moreover, a small change in the relative orientation of additional groups can turn a potent agonist into an antagonist as observed with the potent group III agonist ACPT-I and its stereoisomer ACPT-II endowed with antagonist property (33) (Fig. 2a). We therefore examined whether such small molecular changes of the agonists would turn them into antagonists by possibly preventing the closure of the mGlu VFTM.
Fig 2.
(a) Molecular structures of ligands. (b) 3D model of mGlu8-wt VFTM in its closed form with docked glutamate (black) and model of the D309A mutant with docked ACPT-II (cyan). C-α traces of both models have been superimposed. The distance between central carbon atom of C4-carboxylate of ACPT-II and that of Asp-309 is 3.0 Å, which results in repulsive ionic interaction in the wt. This repulsion is no longer present in mGlu8-D309A. (c) 3D model of mGlu8-wt in its closed form with docked L-AP4 and 3D model of Y227A mutant with docked MAP4. Similar superimposition between wt and Y227A models has been achieved as in b, showing a steric hindrance between the α-methyl group of MAP4 and Tyr-227, which is abolished in mGlu8-Y227A.
Modeling Antagonist Binding in mGlu8 VFTM.
To examine the possible mechanism of action of MAP4 and ACPT-II on mGlu8 receptor, we studied their possible binding mode in a 3D model of the mGlu8 VFTM. This suppress model was generated by comparative modeling based on the coordinates of the mGlu1 VFTM in its closed conformation (PDB ID code :A). Glutamate and L-AP4 then were positioned manually as glutamate in the mGlu1 template, and the resulting models were submitted to energy minimization as described in Materials and Methods. The glutamate and L-AP4 entity of ACPT-II and MAP4, taken in their putative binding conformations, then were respectively superposed to glutamate and L-AP4 in the models. The resulting models (Fig. 2 b and c) show that, in both cases, the closed form of the mGlu8 VFTM cannot easily accommodate the antagonist. In the case of MAP4, the α-methyl group makes steric bumps with the aromatic ring of Tyr-227, whereas the third carboxylic group of ACPT-II, lying close to the carboxylic group of Asp-309, causes ionic repulsion. In contrast, when the same approach was used to dock the mGlu8 potent agonist ACPT-I, a stereoisomer of ACPT-II, the third COO− group was oriented differently at the binding site and can be accommodated well in a closed form of the binding pocket (6, 20, 34).
These data suggest that ACPT-II and MAP4 prevent the formation of a closed form of the mGlu8 VFTM because of ionic and steric hindrance with lobe II, respectively. If the prevention of the formation of a closed state of the VFTM is the main action of these antagonists, one would predict that the removal of these hindrances from lobe II may be sufficient to allow these two compounds to activate the receptor. Indeed, molecular models of a closed form of mutant mGlu8 VFTMs in which Asp-309 or Tyr-227 have been replaced by Ala display a binding site in which putative binding conformations previously mentioned for ACPT-II and MAP4 can fit, respectively (Fig. 2 b and c). When ACPT-II is docked in the D309A mutant, its glutamate moiety is bound as in the wt; moreover, the additional carboxylate (on C4) is now well tolerated in the closed conformation of the VFTM. Similarly, docking of MAP4 in the closed form of Y227A mutant reveals that the glutamate functional groups are bound as in the wt whereas no steric hindrance occurs between the α-methyl substituent and Ala-227.
ACPT-II Is a Full Agonist at mGlu8-D309A Mutant.
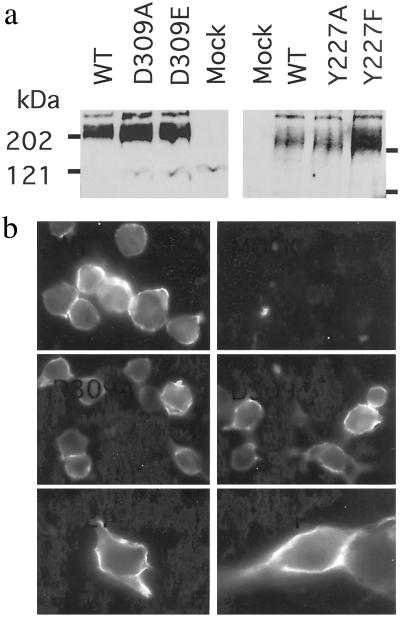
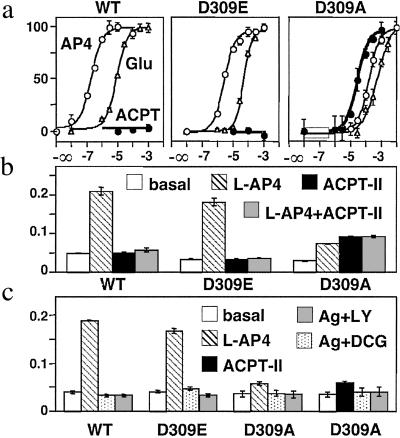
We first examined the effect of mutating Asp-309 from the lobe II of the mGlu8 VFTM into either Ala or Glu on the action of the competitive antagonist ACPT-II. As shown in Fig. 3a, the mGlu8-D309A and -D309E mutants are expressed at a level similar to the wt receptor. Moreover, a similar labeling of cells expressing the wt and mutant receptors tagged at their N-terminal extracellular end was observed on intact cells (Fig. 3b), indicating that they are all correctly targeted to the cell surface. These mutations do not prevent the activation of the receptor by the prototypical group III mGlu receptor agonists L-AP4 or glutamate, although a shift to the right of the agonist dose–response curves is observed by a factor of 4–20 and 500 with mGlu8-D309E and -D309A mutants, respectively (Table 1 and Fig. 4a). A decrease in the maximal response of the agonists also is observed in cells expressing the D309A mutant (Fig. 4b). ACPT-II was found to activate the D309A mutant with an EC50 of 40 μM and a maximal effect equal to that of L-AP4 (Table 1 and Fig. 4a), whereas it antagonizes the action of L-AP4 on both the wt and D309E mutant receptors (Fig. 4b). The agonist activity of ACPT-II on the D309A mutant could be inhibited by the potent group III competitive antagonists LY341495 and DCG-IV (Fig. 4c), further demonstrating that the effect of ACPT-II results from the activation of mGlu8-D309A.
Fig 3.
Expression and surface targeting of the wt, D309A, D309E, Y227F, and Y227A mGlu8 receptors. (a) Western blot analysis of the wild-type and mutant mGlu8a receptor showing the expected molecular mass for their dimeric form. (b) Cells expressing the indicated mGlu8a receptor epitope-tagged at their N-terminal extracellular end were labeled with the HA antibody. Cells were not permeabilized such that only the surface receptor could be detected.
Table 1.
Maximal effects and EC50 values of the tested compounds on mGlu8 receptors and its mutants
| Compound | mGlu8-wt | mGlu8-D309E | mGlu8-D309A | mGlu8-Y227F | mGlu8-Y227A |
|---|---|---|---|---|---|
| Maximal, %AP4 | |||||
| Glu | 96 ± 3 (7) | 100 ± 4 (7) | 68 ± 9 (8) | 90 ± 3 (7) | 81 ± 2 (8) |
| L-AP4 | 100 | 100 | 100 | 100 | 100 |
| ACPT-II | Ant (6) | Ant (4) | 99 ± 18 (8) | n.t. | n.e. (1) |
| MAP4 | Ant (5) | n.e. (1) | n.e. (1) | 26 ± 4 (8) | 85 ± 4 (6) |
| EC50, μM | |||||
| Glu | 15 ± 4 (5) | 66 ± 9 (4) | 790 ± 190 (3) | 28 ± 3 (2) | 103 ± 6 (2) |
| L-AP4 | 0.2 ± 0.1 (3) | 4.1 ± 0.8 (4) | 124 ± 14 (2) | 3.7 ± 1.2 (2) | 5.2 ± 1.2 (2) |
| ACPT-II | (Ki 50) | ND | 40 ± 6 (2) | — | — |
| MAP4 | (Ki 53) | — | — | 25 ± 13 (2) | 11 ± 1 (2) |
The maximal IP production measured with 1 mM l-glutamate, L-AP4, ACPT-II, or MAP4 in cells expressing the indicated receptor is expressed as the percentage of that measured with 1 mM L-AP4 and is mean ± SEM of (n) independent experiments performed in triplicate. The 1 mM L-AP4 effect measured on mGlu8-D309E, -D309A, Y227F, and Y227A is equal to 109 ± 15, 30 ± 3, 180 ± 22, and 153 ± 10% of that measured on control cells (n = 7, 11, 5, and 4, respectively). The EC50 values shown were determined as described in the text and are means ± SEM of n independent determinations. n.t., not tested; n.e., no agonist effect; Ant, antagonist property; ND, not determined. Ki values were calculated based on published data (34).
Fig 4.
ACPT-II is an agonist of mGlu8-D309A receptor. (a) Effect of increasing concentration of L-AP4 (○), glutamate (▵), or ACPT-II (•) on wt, D309E, and D309A mGlu8 receptors. Data are expressed as the percentage of the maximal effect of L-AP4. (b) ACPT-II is an antagonist at both wt and D309E mGlu8 receptors. IP production in cells expressing the indicated receptor under basal (open bars), L-AP4 (hatched bars), 1 mM ACPT-II (solid bars), or L-AP4 and 3 mM ACPT-II (shaded bars). L-AP4 concentrations used were 1.5 μM (wt), 10 μM (D309E), and 300 μM (D309A). Data are expressed as the IP production over the radioactivity remaining in the membranes. (c) Antagonist properties of DCG-IV and LY341495 (both at 1 mM) on wt, D309E, and D309A mGlu8 receptors. Values represent the IP production in cells expressing the indicated receptor under basal (open bars), L-AP4 (hatched bars), 30 μM ACPT-II (solid bars), the agonist plus DCG-IV (dotted bars), and the agonist plus LY341495 (shaded bars). L-AP4 concentrations used were 1.5 μM (wt), 10 μM (D309E), and 100 μM (D309A). Data are means ± SEM of triplicate determinations from typical experiments.
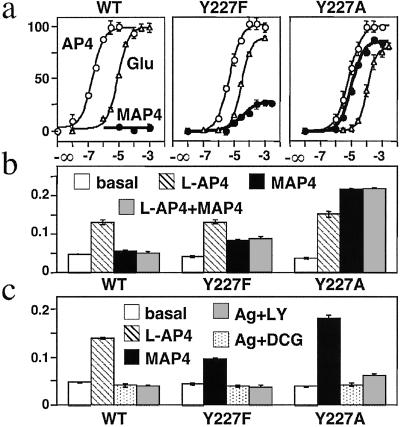
MAP4 Is a Full Agonist at mGlu8-Y227A Mutant.
We then examined the effect of MAP4 on the mGlu8 receptor in which Tyr-227 was replaced by Phe or Ala. As shown in Fig. 3, both mGlu8-Y227F and Y227A are expressed and found at the cell surface. These two mutant receptors still can be activated by the mGlu8 agonists L-AP4 and glutamate, although a slight decrease of their potency was observed with both mutants [EC50 values increased by a factor of 2–10 and 10–20, respectively (Table 1 and Fig. 5a)]. On mGlu8-Y227F, MAP4 behaved as a partial agonist, with an EC50 of 2.9 μM and a maximal effect equal to 26% of that measured with L-AP4 (Table 1 and Fig. 5a). In agreement with a partial agonist action, MAP4 was able to inhibit the effect obtained with a submaximal concentration of L-AP4 (Fig. 5b). On the Y227A mutant, MAP4 was found to be a full agonist, with an EC50 of 11 μM, a value very close to that obtained with L-AP4 (5.2 μM) (Table 1). On either Y227A or Y227F mutants, the effect of MAP4 could be inhibited by the potent mGlu8 competitive antagonists LY341495 and DCG-IV (Fig. 5c).
Fig 5.
MAP4 is a partial agonist and a full agonist of mGlu8-Y227F and mGlu8-Y227A receptors, respectively. (a) Effect of increasing concentration of L-AP4 (○), glutamate (▵), or MAP4 (•) on wt, Y227F, and Y27A mGlu8 receptors. Data are expressed as the percentage of the maximal effect of L-AP4. (b) MAP4 is an antagonist at wt mGlu8 receptor and a partial agonist at mGlu8-Y227F receptor. IP production in cells expressing the indicated receptor under basal (open bars), L-AP4 (hatched bars), 3 mM MAP4 (solid bars), or L-AP4 and 3 mM MAP4 (shaded bars). L-AP4 concentration was 1.5 μM (wt) or 10 μM (Y227F and Y227A). Data are expressed as the IP production over the radioactivity remaining in the membranes. (c) Antagonist properties of DCG-IV (1 mM) and LY341495 (100 μM) on wt, Y227F, and Y227A mGlu8 receptors. Values represent the IP production in cells expressing the indicated receptor under basal (open bars), 1.5 μM L-AP4 (hatched bars), 100 μM MAP4 (solid bars), the agonist plus DCG-IV (dotted bars), and the agonist plus LY341495 (shaded bars). Data are means ± SEM of triplicate determinations from typical experiments.
Discussion
In the present study, we identified two residues in lobe II of the mGlu8 VFTM that likely prevent the formation of a fully closed state in the presence of the antagonists ACPT-II or MAP4, respectively. The mutation into Ala of these residues was sufficient to allow these antagonists to fully activate the mutated receptors. This is consistent with the proposal that a closed form of the mGlu VFTM is required for receptor activation.
In most cases, the interaction of a natural ligand in VFTMs stabilizes a closed state of these bilobate structures, as observed with the bacterial periplasmic-binding proteins (35, 36), the amide-binding protein AmiC (37, 38), and the glutamate-binding domain of both ionotropic (39, 40) and metabotropic receptors (11, 19). In several cases, inhibitors or antagonists have been shown to bind within the cleft that separates both lobes but prevent the full closure of the VFTM. This is the case for the AmiC inhibitor butyramide (38), the ionotropic glutamate receptor 2 (GluR2) antagonist CNQX (39, 40), and, as recently reported, for the mGlu1 antagonist MCPG (19). Such a prevention of the full closure of mGlu VFTMs by antagonists is expected for other large compounds such as the xanthene derivatives of glutamate analogs (LY341495, for example) that obviously cannot fit into the binding pocket of the closed state. Our present data on mGlu8 receptor are consistent with the extension of this antagonist mode of action to smaller antagonists such as MAP4 and ACPT-II, a diastereoisomer of the potent agonist ACPT-I. Indeed, both compounds perfectly adapt to lobe I of the glutamate-binding site; however, the α-methyl group of MAP4 is not accepted in the closed form because of steric hindrance with Tyr-227 from lobe II, and the third carboxylic group of ACPT-II facing Asp-309, also from lobe II, also is not admitted.
From structural studies with a purified mGlu1 VFTM, this domain was shown to dimerize via its lobe I, and the relative orientation of the two modules differs in the glutamate-bound form compared with an empty or MCPG-bound form, as depicted in Fig. 1. However, whether the VFTM closure, the change in the relative orientation of the VFTMs, or both are required for receptor activation is still not known. Moreover, such changes in conformation may not necessarily occur in the full receptor. Our study reveals that mutations of the residues that appear to prevent the mGlu8 VFTM closure on MAP4 or ACPT-II binding can convert these antagonists into full agonists. Indeed, the removal of the negative charge in the D309A mutant allowed ACPT-II to activate the receptor, whereas the conservation of the charge (D309E) did not change the property of this antagonist. In the case of MAP4, the removal of the steric hindrance in the Y227A mutant converts it into a full agonist with a potency very close to that of its analog L-AP4 (EC50 = 5.2 and 10.6 μM, respectively). This indicates that the α-methyl group no longer affects MAP4 interaction in the active form of this mutant receptor.
The removal of the hydroxyl group of Tyr (in the Y227F mutant) already is sufficient to allow partial activation of the receptor by MAP4. In our 3D model, not only is the hydroxyl group in steric clash with the α-methyl group of MAP4, but it also stabilizes the position of the Tyr side chain by means of H bonds with Asp-309, Ser-308, and the backbone of Ser-310. Accordingly, mGlu8-Y227F may be able to accommodate the α-methyl group of MAP4 in a closed form for two reasons: (i) Phe generates less steric hindrance than Tyr, and (ii) the higher mobility of the phenyl group should allow it to adopt a conformation leaving enough space for MAP4. In agreement with this proposal, docking of MAP4 in the wt and Y227F closed binding sites followed by energy minimization revealed a movement of both the phenyl and the hydroxyphenyl rings to accommodate MAP4. However, the H bond network that stabilizes Tyr-227 in the wt receptor is partly lost (data not shown). This makes the movement of the Tyr side chain less likely to occur than that of Phe, explaining why MAP4 can activate the Y227F mutant but not the wt receptor.
In some cases, the angle between the two lobes in the VFTM closed state appears critical for function. For the histidine-binding protein HisJ, the angle of the closed state appears to depend on the ligand and is related to the ability and efficacy of this periplasmic protein to transport the ligand (41). For GluR2, the full VFTM closure is critical for agonist activation and rapid desensitization of the receptor (39, 40). Indeed, kainate, a partial agonist of GluR2, stabilizes a partially closed state of the VFTM (40). Because MAP4 differs from the full agonist L-AP4 only by the presence of the α-methyl group, one expects an accurate full closure to be reached during mGlu8 receptor activation. It is tempting to propose that the fully closed state is not reached with MAP4 in the mGlu8-Y227F mutant, making it a partial rather than a full agonist.
The presently mutated Tyr and Asp residues are part of the conserved motif of all mGlu receptors interacting with the α-amino acid moiety of the ligand (20). As observed in the closed state of the mGlu1 VFTM with bound glutamate (11) and in 3D models for other mGlu receptors (6, 8, 10, 20), Asp makes an ionic interaction whereas Tyr makes a cation–π interaction with the α-amino group. Because these two residues are located in lobe II, they likely play a role in the stabilization of the active liganded closed state. In agreement with this proposal, the mutation of these residues into Ala in the mGlu8 receptor decreased the potency of both L-AP4 and Glu, with the most important effect observed with the mutation of Asp. This suggests that the ionic interaction plays a more important role in the agonist effect than the cation–π interaction and/or that Asp is more critical for the hydrogen bond network mentioned above.
Taken together, our data are consistent with the necessity of the closure of at least one VFTM in the mGlu8 receptor for receptor activation. Does it mean that the change in the relative orientation between the two modules is not required? Aside from the reported structures of the empty, glutamate, and MCPG-bound forms of the dimer of mGlu1 VFTMs, there is no direct demonstration that this occurs in the full receptor. However, the proposal that the change in the relative orientation of the VFTMs, by bringing together their C-terminal ends, plays a role in receptor activation appears an excellent model to explain the original features of the γ-aminobutyric acid type B receptor (21, 22, 42). If this is the case, then the closure of at least one VFTM may be necessary to allow the dimer of VFTMs to reach its “active” orientation. In fact, in the active state, the VFTMs contact each other at the level not only of their lobe I but also of their lobe II (these are quite distant in the “resting” orientation; Fig. 1). Our modeling studies, in agreement with the structures reported recently (19), revealed that the contact area between lobes II in the active orientation of the dimer depends on the open or closed state of each module. Indeed, if this contact area appears to stabilize the active orientation when one VFTM is closed, it is not the case when both VFTMs are open. In that case, clusters of acidic residues are facing each other at the level of lobe II, resulting in an unstable conformation and suggesting that at least one VFTM has to be in the closed state to reach the active orientation. If this hypothesis is correct, mutations of the lobe II interface acidic residues in the open/open active conformation may stabilize it, allowing constitutive activation of the receptor. Such a mutant with constitutive activity has been reported recently (43), consistent with this model for mGlu receptor activation.
Acknowledgments
We thank Julie Kniazeff and Marie-Laure Parmentier for constant support and comments on the manuscript, Hugues-Olivier Bertrand (Accelrys, Orsay, France) for his advice on molecular modeling, and Bernard Mouillac for critical reading of the manuscript. This work was supported by grants from the Centre National de la Recherche Scientifique, the Action Incitative “Physique et Chimie du Vivant” (PCV00–134) from the Centre National de la Recherche Scientifique, the program “Molécules et Cibles Thérapeutiques” from Institut National de la Santé et de la Recherche Médicale and Centre National de la Recherche Scientifique, the foundation Retina France, the Fondation de France (comité Parkinson), and the Action Coordonnée Incitative “Molécules et Cibles Thérapeutiques” from the French government. A.-S.B. was supported by a fellowship from the Fondation de la Recherche Médicale.
Abbreviations
ACPT-I, (1R,3S,4R)-1-aminocyclopentane-1,3,4-tricarboxylic acid
ACPT-II, (1R,3R,4S)-1-aminocyclopentane-1,3,4-tricarboxylic acid
DCG-IV, (2S,2′R,3′R)-2-(2′,3′-dicarboxycyclopropyl) glycine
GPCR, G protein-coupled receptor
HD, heptahelical domain
L-AP4, (S)-2-amino-4-phosphonobutanoic acid
LY341495, (αS)-α-amino-α-[(1S,2S)-2-carboxycyclopropyl]-9H-xanthine-9-propanoic acid
MAP4, (S)-2-amino-2-methyl-4-phosphonobutanoic acid
MCPG, α-methyl-4-carboxyphenylglycine
mGlu, metabotropic glutamate
VFTM, Venus flytrap module
3D, three-dimensional
wt, wild type
HA, hemagglutinin
IP, inositol phosphate
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Bockaert J. & Pin, J.-P. (1999) EMBO J. 18, 1723-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Blasi A., Conn, P. J., Pin, J.-P. & Nicoletti, F. (2001) Trends Pharmacol. Sci. 22, 114-120. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K., Tsuchida, K., Tanabe, Y., Masu, M. & Nakanishi, S. (1993) J. Biol. Chem. 268, 19341-19345. [PubMed] [Google Scholar]
- 4.O'Hara P. J., Sheppard, P. O., Thøgersen, H., Venezia, D., Haldeman, B. A., McGrane, V., Houamed, K. M., Thomsen, C., Gilbert, T. L. & Mulvihill, E. R. (1993) Neuron 11, 41-52. [DOI] [PubMed] [Google Scholar]
- 5.Costantino G. & Pellicciari, R. (1996) J. Med. Chem. 39, 3998-4006. [DOI] [PubMed] [Google Scholar]
- 6.Bessis A.-S., Bertrand, H.-O., Galvez, T., De Colle, C., Pin, J.-P. & Acher, F. (2000) Protein Sci. 9, 2200-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hampson D. R., Huang, X. P., Pekhletski, R., Peltekova, V., Hornby, G., Thomsen, C. & Thogersen, H. (1999) J. Biol. Chem. 274, 33488-33495. [DOI] [PubMed] [Google Scholar]
- 8.Rosemond E., Peltekova, V., Naples, M., Thogersen, H. & Hampson, D. R. (2002) J. Biol. Chem. 277, 7333-7340. [DOI] [PubMed] [Google Scholar]
- 9.Galvez T., Prézeau, L., Milioti, G., Franek, M., Joly, C., Froestl, W., Bettler, B., Bertrand, H.-O., Blahos, J. & Pin, J.-P. (2000) J. Biol. Chem. 275, 41166-41174. [DOI] [PubMed] [Google Scholar]
- 10.Malherbe P., Knoflach, F., Broger, C., Ohresser, S., Kratzeisen, C., Adam, G., Stadler, H., Kemp, J. A. & Mutel, V. (2001) Mol. Pharmacol. 60, 944-954. [DOI] [PubMed] [Google Scholar]
- 11.Kunishima N., Shimada, Y., Tsuji, Y., Sato, T., Yamamoto, M., Kumasaka, T., Nakanishi, S., Jingami, H. & Morikawa, K. (2000) Nature (London) 407, 971-977. [DOI] [PubMed] [Google Scholar]
- 12.Romano C., Yang, W.-L. & O'Malley, K. L. (1996) J. Biol. Chem. 271, 28612-28616. [DOI] [PubMed] [Google Scholar]
- 13.Ray K., Hauschild, B. C., Steinbach, P. J., Goldsmith, P. K., Hauache, O. & Spiegel, A. M. (1999) J. Biol. Chem. 274, 27642-27650. [DOI] [PubMed] [Google Scholar]
- 14.Tsuji Y., Shimada, Y., Takeshita, T., Kajimura, N., Nomura, S., Sekiyama, N., Otomo, J., Usukura, J., Nakanishi, S. & Jingami, H. (2000) J. Biol. Chem. 275, 28144-28151. [DOI] [PubMed] [Google Scholar]
- 15.Romano C., Miller, J. K., Hyrc, K., Dikranian, S., Mennerick, S., Takeuchi, Y., Goldberg, M. P. & O'Malley, K. L. (2001) Mol. Pharmacol. 59, 46-53. [PubMed] [Google Scholar]
- 16.White J. H., Wise, A., Main, M. J., Green, A., Fraser, N. J., Disney, G. H., Barnes, A. A., Emson, P., Foord, S. M. & Marshall, F. H. (1998) Nature (London) 396, 679-682. [DOI] [PubMed] [Google Scholar]
- 17.Jones K. A., Borowsky, B., Tamm, J. A., Craig, D. A., Durkin, M. M., Dai, M., Yao, W.-J., Johnson, M., Gunwaldsen, C., Huang, L.-Y., et al. (1998) Nature (London) 396, 674-679. [DOI] [PubMed] [Google Scholar]
- 18.Kaupmann K., Malitschek, B., Schuler, V., Heid, J., Froestl, W., Beck, P., Mosbacher, J., Bischoff, S., Kulik, A., Shigemoto, R., et al. (1998) Nature (London) 396, 683-687. [DOI] [PubMed] [Google Scholar]
- 19.Tsuchiya D., Kunishima, N., Kamiya, N., Jingami, H. & Morikawa, K. (2002) Proc. Natl. Acad. Sci. USA 99, 2660-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bertrand, H.-O., Bessis, A.-S., Pin, J.-P. & Acher, F. C. (2002) J. Med. Chem., in press. [DOI] [PubMed]
- 21.Galvez T., Duthey, B., Kniazeff, J., Blahos, J., Rovelli, G., Bettler, B., Prézeau, L. & Pin, J.-P. (2001) EMBO J. 20, 2152-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Margeta-Mitrovic M., Jan, Y. N. & Jan, L. Y. (2001) Proc. Natl. Acad. Sci. USA 98, 14643-14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He X.-l., Chow, D.-c., Martick, M. M. & Garcia, K. C. (2001) Science 293, 1657-1662. [DOI] [PubMed] [Google Scholar]
- 24.van den Akker F. (2001) J. Mol. Biol. 311, 923-937. [DOI] [PubMed] [Google Scholar]
- 25.Parmentier M.-L., Prézeau, L., Bockaert, J. & Pin, J.-P. (2002) Trends Pharmacol. Sci. 23, 268-274. [DOI] [PubMed] [Google Scholar]
- 26.Bessis A.-S., Jullian, N., Coudert, E., Pin, J.-P. & Acher, F. (1999) Neuropharmacology 38, 1543-1551. [DOI] [PubMed] [Google Scholar]
- 27.Ango F., Albani-Torregrossa, S., Joly, C., Robbe, D., Michel, J.-M., Pin, J.-P., Bockaert, J. & Fagni, L. (1999) Neuropharmacology 38, 793-803. [DOI] [PubMed] [Google Scholar]
- 28.Brabet I., Parmentier, M.-L., De Colle, C., Bockaert, J., Acher, F. & Pin, J.-P. (1998) Neuropharmacology 37, 1043-1051. [DOI] [PubMed] [Google Scholar]
- 29.Conklin B. R., Farfel, Z., Lustig, K. D., Julius, D. & Bourne, H. R. (1993) Nature (London) 363, 274-276. [DOI] [PubMed] [Google Scholar]
- 30.Gomeza J., Mary, S., Brabet, I., Parmentier, M.-L., Restituito, S., Bockaert, J. & Pin, J.-P. (1996) Mol. Pharmacol. 50, 923-930. [PubMed] [Google Scholar]
- 31.Parmentier M. L., Joly, C., Restituito, S., Bockaert, J., Grau, Y. & Pin, J.-P. (1998) Mol. Pharmacol. 53, 778-786. [PubMed] [Google Scholar]
- 32.Blahos J., Fischer, T., Brabet, I., Stauffer, D., Rovelli, G., Bockaert, J. & Pin, J.-P. (2001) J. Biol. Chem. 276, 3262-3269. [DOI] [PubMed] [Google Scholar]
- 33.Acher F., Tellier, F., Azerad, R., Brabet, I., Fagni, L. & Pin, J.-P. (1997) J. Med. Chem. 40, 3119-3129. [DOI] [PubMed] [Google Scholar]
- 34.De Colle C., Bessis, A.-S., Bockaert, J., Acher, F. & Pin, J.-P. (2000) Eur. J. Pharmacol. 394, 17-26. [DOI] [PubMed] [Google Scholar]
- 35.Quiocho F. A. (1990) Philos. Trans. R. Soc. London B 326, 341-351. [DOI] [PubMed] [Google Scholar]
- 36.Wolf A., Shaw, E. W., Nikaido, K. & Ames, G. F.-L. (1994) J. Biol. Chem. 269, 23051-23058. [PubMed] [Google Scholar]
- 37.Pearl L., O'Hara, B., Drew, R. & Wilson, S. (1994) EMBO J. 13, 5810-5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Hara B. P., Wilson, S. A., Lee, A. W., Roe, S. M., Siligardi, G., Drew, R. E. & Pearl, L. H. (2000) Protein Eng. 13, 129-132. [DOI] [PubMed] [Google Scholar]
- 39.Armstrong N., Sun, Y., Chen, G. Q. & Gouaux, E. (1998) Nature (London) 395, 913-917. [DOI] [PubMed] [Google Scholar]
- 40.Armstrong N. & Gouaux, E. (2000) Neuron 28, 165-181. [DOI] [PubMed] [Google Scholar]
- 41.Wolf A., Lee, K. C., Kirsch, J. F. & Ames, G. F.-L. (1996) J. Biol. Chem. 271, 21243-21250. [DOI] [PubMed] [Google Scholar]
- 42.Havlickova, M., Prezeau, L., Duthey, B., Bettler, B., Pin, J.-P. & Blahos, J. (2002) Mol. Pharmacol., in press. [DOI] [PubMed]
- 43.Jensen A. A., Sheppard, P. O., Jensen, L. B., O'Hara, P. J. & Bräuner-Osborne, H. (2001) J. Biol. Chem. 276, 10110-10118. [DOI] [PubMed] [Google Scholar]