Abstract
Glycerol kinase from Escherichia coli, but not Haemophilus influenzae, is inhibited allosterically by phosphotransferase system protein IIAGlc. The primary structures of these related kinases contain 501 amino acids, differing at 117. IIAGlc inhibition is transplanted from E. coli glycerol kinase into H. influenzae glycerol kinase by interconverting only 11 of the differences: 8 residues that interact with IIAGlc at the allosteric binding site and 3 residues in the conserved ATPase catalytic core that do not interact with IIAGlc but the solvent accessible surface of which decreases when it binds. The three core residues are crucial for coupling the allosteric site to the conserved catalytic core of the enzyme. The site of the coupling residues identifies a regulatory locus in the sugar kinase/heat shock protein 70/actin superfamily and suggests relations between allosteric regulation and the active site closure that characterizes the family. The location of the coupling residues provides empirical validation of a computational model that predicts a coupling pathway between the IIAGlc-binding site and the active site [Luque, I. & Freire, E. (2000) Proteins Struct. Funct. Genet. Suppl. 4, 63–71]. The requirement for changes in core residues to couple the allosteric and active sites and switching from inhibition to activation by a single amino acid change are consistent with a postulated mechanism for molecular evolution of allosteric regulation.
Allosteric regulation of enzyme catalysis is a fundamental aspect of control in biological systems. For most classically studied allosteric enzymes (1), catalytic activity is modulated by small molecules such as fructose 1,6-bisphosphate (FBP), phosphoenolpyruvate, or nucleotides. The regulatory binding sites and the active sites are located at subunit–subunit interfaces of the homooligomeric proteins and are separated by >10 Å. Substrate binding shows homotropic interactions, typically positive cooperativity. Allosteric effectors change the affinity for substrates at the active sites but do not affect Vmax. Allosteric inhibition decreases apparent affinity for substrate, and activation increases apparent affinity. Changes in quaternary structure, involving small rigid-body rotations and translations of subunits, are seen in crystal structures of the enzymes with substrates or allosteric effectors. Regulation of catalysis by protein–protein interactions is less understood but seems to be the rule rather than the exception, and there is widespread interest in interactomes and roles of macromolecular interactions in living systems (2). A related area of interest is the molecular evolution of allosteric control by protein–protein interactions. Here we show that transplantation of allosteric regulation from one enzyme to another provides insights into how protein–protein interactions distant from an active site regulate catalysis and into mechanisms of molecular evolution.
Glycerol kinase (EC 2.7.1.30; ATP:glycerol 3-phosphotransferase) from Escherichia coli (EcGK) is a regulatory enzyme (3). It is inhibited allosterically by IIAGlc, the 18-kDa glucose-specific phosphocarrier protein of the phosphoenolpyruvate:glycose phosphotransferase system (4). Binding of IIAGlc decreases Vmax and the substrate Michaelis constants, and no cooperativity is observed for inhibition or with respect to glycerol or MgATP (5). Thus, in contrast to classically studied regulatory enzymes, IIAGlc inhibition of EcGK shows decreased Vmax, increased apparent substrate affinity, and no homotropic interactions for any of the ligands. X-ray crystallographic studies of the protein–protein complex show that EcGK is a homotetramer with one molecule of IIAGlc bound per 56-kDa subunit (Fig. 1A; refs. 6 and 7). The allosteric site is on the subunit surface more than 30 Å from the catalytic site and not at a subunit–subunit interface. The structure of the complex elicits the question of how binding of a regulatory protein to the subunit surface distant from the catalytic site produces the regulation of catalysis that is observed for EcGK. However, typical approaches to addressing this question have not yielded a complete answer.
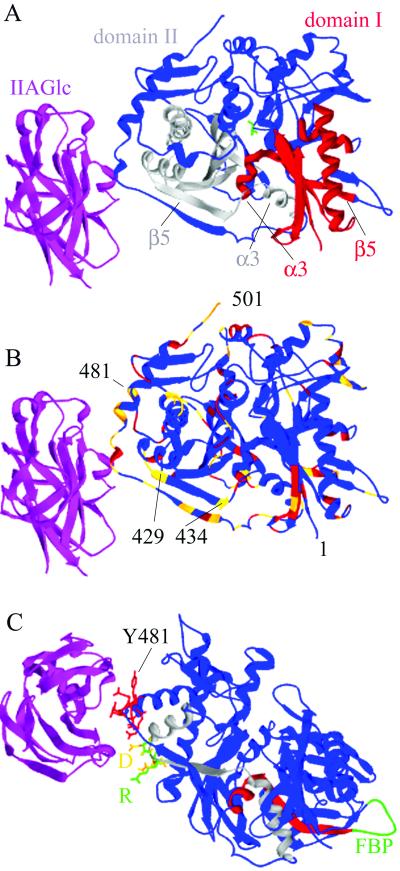
Fig 1.
Ribbon diagrams of the structure of one EcGK subunit with bound IIAGlc. IIAGlc is shown in magenta, and EcGK is shown in the remaining colors. (A) The conserved ATPase catalytic core. Glycerol is shown in green in the active site. Domain II, which contains the IIAGlc binding site, is to the left of the cleft, and domain I is to the right. The conserved ATPase motif in domain I is shown in red and in domain II in gray. The secondary structure elements α3 and β5 are labeled in each domain. (B) Amino acid differences in EcGK and HiGK. Identities are shown in blue, conservative substitutions in red, and nonconservative substitutions in gold. The N- and C-terminal positions are labeled 1 and 501, respectively, and selected residue positions are numbered. (C) The location of amino acid substitutions required to transplant IIAGlc inhibition to HiGK. The amino acids that interact directly with IIAGlc are shown as stick models in red. Amino acids 427–429 in EcGK (GTR) are shown in green and in HiGK (DVN) are shown in gold. D427 is labeled D and R429 is labeled R. Conserved ATPase catalytic core elements are colored as described for A. The loop with the binding site for FBP in domain I is shown in green and labeled FBP. The images were generated from PDB ID code by using SWISS-PDB VIEWER 3.5 (15) and rendered by using POV-RAY 3.1 for WINDOWS (www.povray.org/).
Crystallographic studies do not provide a complete structural description of IIAGlc inhibition. They verify that EcGK is a member of the sugar kinase/heat-shock protein 70/actin superfamily of enzymes, the members of which fulfill many important functions (8). Superfamily members have a conserved ATPase catalytic core, consisting of the motif β1β2β3α1β4α2β5α3 in each domain (Fig. 1A). A hallmark of the superfamily, exemplified by hexokinase in glucose binding, is a conformational change that involves relative motion of two domains (I and II) to close the active site cleft. Thus, the active site is located at a domain–domain interface within a single subunit. The closure has been characterized as a domain-shear motion involving movement of the α3 element in each domain relative to the same element in the other and movement of the core β-sheet in each domain relative to the α3 element of the other (9). Posttranslational regulation of the function of many of the family members is believed to involve modulation of this motion. Crystallographic studies of EcGK show evidence for conformational changes associated with active site closure and induced fit of amino acids that bind IIAGlc but do not show how these two conformational changes are coupled (10, 11). The conformation of IIAGlc does not change after binding to EcGK.
Phylogenetics studies of this family (12) do not provide further insights into the allosteric regulation. Only EcGK and the essentially identical enzymes from Salmonella are known to be inhibited by IIAGlc. Thus, there are not multiple examples of regulated enzymes that show conserved amino acids to identify molecular coupling pathways (13). However, these studies identify sugar kinases related to EcGK. The existence of these related enzymes suggests the use of transplantation of IIAGlc inhibition into a naive enzyme as a gain-of-function approach to identify amino acid differences in EcGK that give rise to IIAGlc allosteric control and to investigate mechanisms of molecular evolution of allosteric control.
Transplantation of allosteric regulation has been used in hemoglobins (14). The molecular basis for bicarbonate-ion control of the oxygen affinity of crocodilian hemoglobin was postulated from primary structure comparisons and tested by site-directed mutagenesis. Bicarbonate-ion control can be transplanted to human hemoglobin by 12 amino acid substitutions to generate a putative bicarbonate-ion binding site at the tetramer interface. Although the ion binding site was not verified structurally, the transplantation indicates that functionally important loci can be identified from amino acid replacements necessary to elicit a new function. Thus, a subset of the amino acids that participate in IIAGlc inhibition of EcGK may be identified as the replacements from EcGK into a naive enzyme that are necessary to transplant inhibition.
A family member closely related to EcGK is glycerol kinase from Haemophilus influenzae (HiGK). It contains 502 amino acid residues, one more than EcGK, and the primary structures are 87% similar (76% identical). Homology modeling (ref. 15; www.expasy.ch/) of the HiGK sequence to the known structure of EcGK gives overall root-mean-square deviation of the backbone atoms of 0.43 Å, consistent with the high degree of similarity. Locations of the 117 amino acid differences are shown in Fig. 1B. We purified HiGK and characterized its catalytic and regulatory properties (16). Both EcGK and HiGK are inhibited allosterically by FBP. Crystallographic studies of EcGK show two FBP-binding sites per tetramer located at subunit–subunit interfaces (17). The binding site for FBP on EcGK is located in domain I on the opposite side of the subunit from the binding site for IIAGlc, and the FBP-binding amino acid residues are conserved in the H. influenzae enzyme (Fig. 1C). In solution, EcGK shows an equilibrium between homodimers and homotetramers (18), and FBP inhibition requires tetramer formation (19). Sedimentation velocity ultracentrifugation shows that HiGK sediments more slowly than EcGK in the absence of FBP, suggesting the presence of a greater fraction of dimer but about the same as EcGK in the presence of FBP. The catalytic, structural, and FBP regulatory behavior of HiGK is similar to EcGK, but IIAGlc regulation is different. Eight of the eleven EcGK amino acids that interact with IIAGlc are different in HiGK, and it is not inhibited by E. coli IIAGlc. This lack of inhibition could indicate complementary substitutions in H. influenzae IIAGlc, but its sequence is highly similar to the E. coli IIAGlc (82% similarity/72% identity), and all the amino acids that interact with EcGK (7) are identical in H. influenzae IIAGlc. Although it has not been tested, inhibition of HiGK by its own IIAGlc is unlikely, and roles for IIAGlc in the regulation of glycerol metabolism in H. influenzae have not been described. Allosteric inhibition of EcGK by FBP and IIAGlc operate independently, and IIAGlc inhibition does not require tetramer formation (19–21). Thus, HiGK is naive with respect to allosteric control by E. coli IIAGlc.
In the simplest case, replacement of only the eight different interacting amino acid residues in HiGK with those from EcGK would be sufficient to transplant IIAGlc inhibition. However, we show that this is not the case. Transplantation of IIAGlc inhibition into HiGK requires substitution of three amino acid residues of the conserved ATPase catalytic core, and IIAGlc inhibition of EcGK can be switched to activation by changing one of these residues. The replacement of amino acids in the conserved core to couple the IIAGlc binding site to the catalytic site and the switching behavior provide experimental verification of a postulated mechanism for evolution of allosteric enzyme regulation (22, 23). The coupling locus is identical topologically to the FBP-binding site in domain I and is consistent with a molecular pathway that is predicted by a structure-based computational method to couple IIAGlc binding and the active site (24), suggesting a relation between the proposed domain-shear motion and molecular mechanisms for allosteric regulation of superfamily members. The coupling amino acid residues in EcGK do not interact with IIAGlc but do undergo changes in solvent-accessible surface area after complex formation, suggesting that such changes play crucial roles in protein–protein regulatory interactions.
Methods
Materials.
General reagents, phosphoenolpyruvate, NADH, pyruvate kinase, and lactate dehydrogenase were purchased from Sigma. Restriction enzymes and T4 DNA ligase were purchased from Promega. IIAGlc was generously supplied by Saul Roseman and Norman D. Meadow of the Department of Biology at The Johns Hopkins University (Baltimore).
Construction of Site-Directed Variants and Chimera.
The plasmids harboring the genes for the chimeric enzymes H:E245 and E:HII245 were constructed by ligation of DNA fragments generated from EcoRI, HindIII, and BstEII restriction endonuclease digestion of the plasmids harboring the genes for the parent enzymes.‡ Plasmids harboring the genes for the chimeric enzymes HII:E353–450, HII:E423–442, and revertants of HII:E423–442 were constructed following the ExSite protocol of Stratagene. The remaining plasmids harboring genes for chimeric enzymes were constructed by using overlap extension PCR (25). The coding regions for all chimeric enzymes were sequenced and found to possess the expected sequences.
Protein Purification.
HiGK was purified as described (16). All other glycerol kinases were purified essentially as described (21), with the following changes: only the first two chromatography steps were needed for H:E245; only the first chromatography step was needed for HiGKII, HiGKII–GTR, and HiGK–GTR, with the modification that samples were loaded in the absence of added salt and eluted by a 0.1 M KCl step gradient; and an additional chromatography step using phenyl-Sepharose (26) was needed for HiGKII–D427G.
Kinetic Assays.
Enzyme assays were performed and analyzed as described (16) by using an ADP-coupled spectrophotometric assay at pH 7.0 and 25°C. A unit is defined as the amount of glycerol kinase that produces 1 μmol of ADP per min in this assay. For determination of KATP, the glycerol concentration was 2 mM; for determination of Kgol, the ATP concentration was 1 mM for H:E245, 0.7 mM for HiGK–GTR, 0.5 mM for HiGK, HiGKII–GTR, and HiGKII–D427G, 0.3 mM for HiGKII, and 0.1 mM for EcGK, EcGK–G427D, and EcGK–DVN.
For IIAGlc inhibition assays, ATP and glycerol concentrations were 2.5 and 2 mM, respectively, IIAGlc concentrations varied from 0 to 27 μM, and 0.1 mM ZnCl2 was added to the assays. Inhibition data were fitted to a normalized hyperbolic function: SA,% = 100 − [100 × (Imax × [IIAGlc])/K50 + [IIAGlc])], where SA,% is the percent remaining activity of the enzyme at a particular inhibitor concentration compared with the activity with no effector present, Imax is the maximal percent inhibition, and K50 is the apparent dissociation constant for effector binding. The data were fitted by using KALEDIAGRAPH 3.09 for WINDOWS (Synergy Software, Reading, PA).
Sedimentation Velocity Ultracentrifugation.
These experiments were performed as described (27) by using a Beckman Model XL-A analytical ultracentrifuge at 35,000 rpm and 25°C. The concentration of glycerol kinase was 5 μM (subunits), and that of IIAGlc, when present, was 35 μM. ZnCl2 was included in all buffers at 10 μM. Data were analyzed and the sedimentation coefficient was determined by using the program SVEDBERG 6.38 by John Philo (28).
Results and Discussion
Substitution of the IIAGlc-Interacting Amino Acids Is Not Sufficient to Transplant IIAGlc Control.
Crystallographic studies identify 11 amino acids of EcGK that interact with IIAGlc: R402 and P472GIETTERNY481 (7). Amino acid 402 is the same in both enzymes, but 8 of the 10 sequentially contiguous amino acids are different (P472DSDNEKRER481, with differences underlined; ref. 16). To determine whether substitution of the interacting EcGK residues into HiGK results in transplantation of IIAGlc inhibition, the chimeric enzyme, HiGKII was constructed (Table 1). The catalytic activity of HiGKII, similar to its HiGK parent, is not altered detectably by IIAGlc. The kinetic parameters and allosteric inhibition by FBP (not shown) are little altered by the substitutions, indicating that they do not perturb the enzyme structure greatly.
Table 1.
Catalytic and regulatory properties of chimeric enzymes
| Enzyme
|
Kinetic parameters | IIAGlc inhibition | |||
|---|---|---|---|---|---|
| Vmax, units/mg | Katp, μM | Kgol, μM | Imax, % | K50, μM | |
| EcGK | 13 ± 1 | 6 ± 1 | 9 ± 1 | 94 ± 1 | 0.5 ± 0.1 |
| HiGK | 21 ± 1 | 45 ± 6 | 34 ± 2 | n.e. | n.e. |
| HiGKII | 9 ± 1 | 24 ± 3 | 34 ± 2 | n.e. | n.e. |
| HiGKII–GTR | 25 ± 2 | 46 ± 8 | 33 ± 2 | 69 ± 1 | 2.0 ± 0.1 |
| H:E245 | 37 ± 1 | 19 ± 1 | 17 ± 1 | 74 ± 1 | 0.7 ± 1 |
| HiGK–GTR | 20 ± 2 | 73 ± 12 | 48 ± 3 | n.e. | n.e. |
| EcGK–DVN | 10 ± 1 | 3 ± 1 | 3 ± 1 | −15 ± 2 | 2 ± 0.9 |
| EcGK–G427D | 9 ± 1 | 3 ± 1 | 3 ± 1 | −14 ± 2 | 6 ± 3 |
| HiGKII–D427G | 23 ± 3 | 69 ± 15 | 62 ± 3 | 80 ± 14 | 29 ± 8 |
Steady-state kinetic and regulatory properties of the purified enzymes were determined as described under Methods.
HiGKII contains substitutions of the amino acids from EcGK in the sequentially contiguous portion of the IIAGlc binding site (positions 472–481): HiGK, PDSDNEKRER; HiGKII, PGIETTERNY. Additional substitutions at positions 427–429 are shown after the hyphen.
n.e., no effect.
Sedimentation velocity ultracentrifugation (as described under Methods) was used to evaluate IIAGlc binding (Table 2). The sedimentation coefficient of HiGKII is the same as that of HiGK, further showing that the substitutions do not perturb the structure greatly. The addition of IIAGlc increases the sedimentation coefficient for EcGK but does not alter the sedimentation coefficient of HiGK or HiGKII, which is consistent with binding of IIAGlc to EcGK but not to the other two enzymes. The detection limits of this method indicate that the affinity for IIAGlc binding to these enzymes is at least 200-fold less than for EcGK. Thus, substitution of the interacting amino acids from EcGK is not sufficient to transplant IIAGlc binding, hence allosteric control.
Table 2.
Binding of IIAGlc to glycerol kinases: Sedimentation velocity ultracentrifugation
| Enzyme
|
Sedimentation coefficient, S | |
|---|---|---|
| −IIAGlc | +IIAGlc | |
| EcGK | 10.2 ± 0.1 | 12.5 ± 0.1 |
| HiGK | 6.7 ± 0.1 | 6.8 ± 0.1 |
| HiGKII | 6.6 ± 0.1 | 6.5 ± 0.1 |
| HiGKII–GTR | 6.8 ± 0.1 | 7.8 ± 0.1 |
Identification of Additional Substitutions That Are Required to Transplant IIAGlc Inhibition.
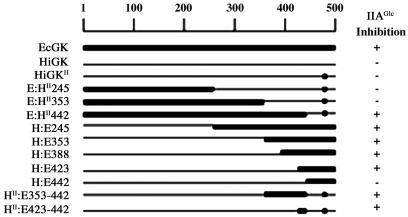
Fig. 1B shows that the 117 primary structure differences between EcGK and HiGK occur in both domains, 69 in domain II, and in the conserved catalytic core and regions outside the core. To identify the adaptive substitutions, a series of chimeric enzymes was constructed to determine portions of the primary structure that must be interchanged to transplant IIAGlc inhibition (Fig. 2). All the chimeric enzymes containing EcGK amino acids 423–442 in addition to the interacting amino acids from EcGK are inhibited by IIAGlc. For those that contain the amino acids from HiGK at positions 423–442, inhibition by IIAGlc could not be detected, i.e., less than 3% inhibition, indicating that the apparent affinity is at least 100-fold less than that for EcGK.
Fig 2.
Identification of EcGK primary structure that transplants IIAGlc inhibition to HiGK. The primary structure of EcGK is represented by the thicker line, and that of HiGK is represented by the thinner line. Chimeric enzymes were constructed as described under Methods. IIAGlc inhibition was assessed by determining the specific activity of glycerol kinase in cell extracts without and with the addition of IIAGlc to the assay. +, IIAGlc inhibits the chimeric enzyme; −, no IIAGlc inhibition observed.
There are six amino acid differences between HiGKII and EcGK in this region: positions 423, 427–429, 434, and 436. The contribution of each of these differences was assessed by using the chimeric enzyme HII:E423–442, which has the amino acids from EcGK at these positions. Each of these amino acids in the chimeric enzyme was reverted individually to the amino acid in HiGK, and the response of the enzyme to IIAGlc was determined (Table 3). The reversions at positions 427–429 show substantial effects on inhibition by IIAGlc, whereas those at the other three positions show little or no effect. The revertant G427D shows activation instead of inhibition. It is important to note that the absence of substantial effects for revertants at the other positions does not indicate that they do not participate in IIAGlc binding and inhibition. The small but significant decrease in inhibition seen for the R436K revertant, a conservative substitution, indicates that this residue indeed has a role in IIAGlc inhibition, but its replacement is not necessary to transplant the inhibition.
Table 3.
Contributions of EcGK amino acids 423–436 to IIAGlc inhibition of chimeric enzyme HII:E423–442
| Amino acid substitution in HII:E423–442 | vi/vo |
|---|---|
| None | 0.50 |
| S423A | 0.50 |
| G427D | 1.12 |
| T428V | 0.38 |
| R429N | 0.22 |
| E434V | 0.52 |
| R436K | 0.46 |
Glycerol kinase activity in crude cellular extracts was determined in duplicate without and with 10 μM IIAGlc added to the assay and corrected for glycerol-independent background rates. IIAGlc inhibition is expressed as the ratio of the reaction velocity with IIAGlc, vi, to the velocity without IIAGlc, vo. Duplicate values for vi/vo differed from the reported average value by 0.01 or less.
Amino acid substitutions in this chimeric enzyme are shown by using the single-letter abbreviations and sequence number, where the amino acid replacement is the residue in HiGK.
Determination of the IIAGlc concentration dependence of inhibition for the HII:E423–442 parent enzyme yielded K50 = 3 ± 0.2 μM and Imax = 65 ± 1%.
Based on the greater sensitivity of IIAGlc inhibition to the reversions at positions 427–429, the variant HiGKII–GTR, in which EcGK amino acids replace the HiGK amino acids at positions 473–481 and 427–429, was constructed. It shows substantial inhibition by IIAGlc, whereas its catalytic properties differ little from those of EcGK, HiGK, or HiGKII (Table 1). The apparent affinity for IIAGlc is 4-fold less and the maximum inhibition by IIAGlc is less than that for EcGK. These parameters agree well with those for the H:E245 chimeric enzyme (Table 1), showing that most of the contribution of the 69 domain II differences to IIAGlc inhibition is obtained with only 11 substitutions. The parameters also agree well with those obtained for the chimeric enzyme HII:E423–442 (Table 3), further supporting the conclusion that substitutions of the other three different amino acids (423, 434, and 436) are not necessary to transplant IIAGlc inhibition. The differences between HiGKII–GTR and HII:E423–442 at these three positions do not account for the lower affinity and extent of inhibition relative to EcGK. Binding of IIAGlc to HiGKII–GTR can be detected in the sedimentation velocity ultracentrifugation assay (Table 2). Thus, IIAGlc binding and regulation of catalysis are highly interdependent. The increase in the sedimentation coefficient for HiGKII–GTR is about one-half that obtained for EcGK. This smaller increase suggests that IIAGlc does not alter the oligomeric state of HiGKII–GTR, which is consistent with the independence of IIAGlc inhibition and dimer–tetramer assembly seen for EcGK (19).
To determine whether the changes at positions 427–429 alone are sufficient to transplant IIAGlc inhibition, the variant HiGK–GTR was constructed. This variant retains the HiGK amino acids at positions 473–481 and is not inhibited by IIAGlc (Table 1). Substitution of HiGK amino acids with those from EcGK at positions 427–429 is necessary but not sufficient for IIAGlc inhibition. Thus, substitution of only 11 of the 117 different amino acids from EcGK into HiGK is necessary and sufficient to transplant substantial IIAGlc inhibition.
The variant EcGK–DVN, in which the amino acids at positions 427–429 in EcGK are replaced with those from HiGK, is the converse of HiGKII–GTR. It was constructed to determine whether these substitutions are sufficient to abolish IIAGlc binding and inhibition as seen for HiGKII, which has DVN at these positions. The EcGK–DVN variant binds IIAGlc, and its apparent affinity is 4-fold less than EcGK, which is about the same as for HiGKII–GTR (Table 1). However, it displays greatly altered behavior with respect to IIAGlc action, which is switched from inhibition to activation. Activation by IIAGlc was observed for the revertant HII:E423–442–G427D (Table 3). This revertant corresponds to the variant EcGK–G427D, in which only amino acid 427 is replaced with the corresponding one from HiGK. The apparent role of amino acid 427 in activation by IIAGlc was verified by constructing the variant EcGK–G427D. This variant shows activation rather than inhibition by IIAGlc, and the apparent affinity for IIAGlc is decreased 11-fold (Table 1). Thus, the converse substitution of HiGK amino acids for EcGK at 427–429 does not abolish IIAGlc binding but switches its effect from inhibition to activation, and the substitution G427D in EcGK is sufficient for the switching.
The variant HiGKII–D427G was made to evaluate the extent to which D427 is responsible for the lack of IIAGlc binding to HiGKII. It retains the amino acids from HiGK at positions 428 and 429 but has the glycine from EcGK at position 427. This variant is inhibited by IIAGlc; however, the apparent affinity for IIAGlc is ≈15-fold less than that of HiGKII–GTR (Table 1). Thus, the single substitution, D427G, in combination with the interacting residues is sufficient for IIAGlc binding and inhibition, but the amino acids at positions 428 and 429 make significant contributions to the affinity for IIAGlc. Contributions of these two positions are seen also in the T428V and R429N revertants of HII:E423–442 (Table 3). The revertants show decreased inhibition, which is consistent with the reduced affinity seen for HiGKII–D427G.
Crystallographic studies show that the conformations of EcGK residues 474–478 undergo induced-fit transitions between coil, 310 helix, and α-helix upon IIAGlc binding (11). Alanine substitutions at positions 479–481, which are α-helical in the absence or presence of IIAGlc, result in large decreases in affinity that are consistent with stabilization of the α-helix, affecting positions that undergo the conformational changes (29). On the basis of the earlier reports, the lack of detectable IIAGlc binding to HiGKII strongly suggests that this region is unable to undergo the induced fit, and the IIAGlc binding that is observed after substitution of the amino acids at 427–429 strongly suggests that the conformational change is then possible. Verification of the conformation in this region will require crystallographic studies. Additional conformations may be observed, given the switching from inhibition to activation that is obtained depending on the identity of the amino acid at position 427.
Positions 427–429 Couple IIAGlc Binding to the Conserved ATPase Catalytic Core and Identify a Key Regulatory Locus in Both Domains.
Positions 427–429 are critical for the binding of IIAGlc and coupling to the active site. Crystal structures show that these amino acids do not form interactions with IIAGlc, and there is no change in conformation in this region after IIAGlc binding that is discernible at the 2.4–3.0-Å resolutions of the structures (7, 10, 11). No atoms of the amino acids at 427–429 are closer than 4.3 Å to IIAGlc, and no IIAGlc amino acid side chains form polar interactions with these amino acids. Analysis of accessible surface areas of these amino acids shows that IIAGlc occludes them from solvent (E. Freire, personal communication). Their accessible surface areas in the absence and presence of IIAGlc in the EcGK–IIAGlc complex (PDB ID code , 2.6-Å resolution) were calculated according to the Lee and Richards algorithm by using a probe radius of 1.4 Å and a slice width of 0.25 Å (30). The accessible surface area of the amino acids at positions 427 and 429 is decreased by a total of 47 Å2 (6% of the total buried surface area) in the presence of IIAGlc. Thus, amino acid residues that undergo changes in solvent-accessible surface area but do not make intermolecular contacts can play critical roles in binding and allosteric regulation. This may be an important, previously uncharacterized aspect of protein–protein regulatory interactions relative to small molecule allosteric effectors.
The location of amino acids 427–429 in EcGK provides insights into their critical role in IIAGlc regulation (Fig. 1C). It is in the conserved ATPase catalytic core, connecting the elements α2 and β5 in domain II. Thus, the substitutions at 427–429 in HiGKII–GTR represent changes in the core necessary for IIAGlc to bind and for coupling the binding to inhibition. These residues are located on the subunit surface at the distal end of element β5, which connects them directly to element α3, an active site α-helix which is proposed to move during the domain-shear motion that closes the active site (9). The experimentally determined importance of amino acids 427–429 in IIAGlc inhibition of EcGK verifies computational studies of Freire and coworkers (24, 31, 32). Their results predict that element β5 of domain II is the primary pathway that couples the IIAGlc binding site to structural stability at the active site, specifically helix α3.
The binding site for FBP in domain I is identical topologically to that for IIAGlc in domain II (Fig. 1C; refs. 7 and 17). Thus, 427–429 and the FBP site identify a regulatory locus that is common to the conserved catalytic core motif in each domain. Regulatory interactions at this locus in each case can result in inhibition or activation. As shown here, if the glycine at 427 in EcGK is replaced by aspartate, activation rather than inhibition is obtained. In the case of the related glycerol kinases from Gram-positive bacteria, phosphorylation at the locus corresponding to the FBP-binding site results in activation of catalysis (33). For both 427–429 and the FBP site, the locus is a junction between the conserved catalytic core and additions or insertions that expand the functions of the core. Positions 427–429 are at a junction between the core and the C-terminal addition subdomain IIC (positions 451–501), which forms a substantial portion of the dimer interface. The FBP site is at a junction between the core and insertion subdomain IC. In each case, the subdomain forms a layer of structure above the layer that is formed by the conserved α1 and α2 elements. In the proposed domain-shear motion, the α1 and α2 elements move with the β-sheet in each domain (9). The importance of interactions between the layers is indicated by the effect of a single amino acid substitution in EcGK on IIAGlc inhibition (20). The substitution G304S alters interactions between the conserved β-sheet and element α1 in domain II and switches the effect of IIAGlc to activation. The location of the common regulatory locus in each domain suggests that the allosteric control is mediated through changes in the motion of the subdomain relative to the conserved core elements. However, structural constraints that arise from the necessity for maintaining the tertiary structure of the subunit preclude large shifts in conformation and different interdigitating side chain patterns between the layers, such that small motions are sufficient for catalytic control. Thus, for this widely distributed protein superfamily, small motions between layers of secondary structure in the subunit seem to have the same roles as the rigid-body rotations and translations of entire subunits observed for classically studied regulatory enzymes.
It is important to note that the transplanted amino acids identify only differences between EcGK and HiGK that have to be changed to obtain IIAGlc inhibition. Doubtless, several of the many identical and conserved amino acid positions also are important for allosteric control. In particular, elements of the conserved catalytic core must have roles. The transplanted amino acids thus identify only a subset of the positions that are involved in allosteric regulation of glycerol kinases by IIAGlc. However, they represent a key subset in the evolution of that allosteric control that is necessary for generating the allosteric binding site and coupling it to the conserved catalytic core.
Molecular Evolution of Allosteric Regulation.
A mechanism for molecular evolution of allosteric regulation was postulated on the basis of structural and phylogenetic studies of glycogen phosphorylases (22, 23). Acquisition of 5′-AMP activation of vertebrate glycogen phosphorylase is thought to require amino acid substitutions at a preexisting site for the allosteric inhibitor glucose 6-phosphate to generate a site for 5′-AMP. In addition, substitutions to modify interactions between secondary structural elements of the conserved catalytic core generate a “switch” that allows 5′-AMP binding to activate, and glucose 6-phosphate binding to inhibit, catalysis at the distant active site. This core-coupling mechanism for acquisition of 5′-AMP activation of vertebrate glycogen phosphorylase has not been tested by gain-of-function studies. Furthermore, the proposal involves modification of an existing regulatory mechanism rather than acquisition of regulation by a naive enzyme.
The binding site for IIAGlc on EcGK is primarily on a C-terminal addition onto the last α3 element of the conserved motif of domain II, termed subdomain IIC (amino acids 451–501; ref. 7). The portion of the addition that is located C-terminally to the IIAGlc binding site is a long α-helix that forms a substantial portion of the subunit–subunit interface in the dimer. The three members of the sugar kinase family that have been characterized, EcGK (18), HiGK (16), and E. coli l-ribulokinase (34), form dimers, but only EcGK is inhibited by IIAGlc (16). Molecular evolution of these family members thus may have involved a gene-fusion event to give a C-terminal addition to the conserved ATPase core and allow dimer formation. Results presented here show that a few amino acid substitutions in the conserved core can couple binding of IIAGlc on this addition to modulation of catalysis at the active site and a single amino acid change is sufficient to switch the modulation from inhibition to activation with only modest effects on binding affinity. The experimental results from gain-of-function studies reported here thus are consistent with the postulated mechanism for molecular evolution of allosteric regulation. The same mechanism seems, as expected, to account for molecular evolution of allosteric regulation by both small molecules and protein–protein interactions.
Acknowledgments
We thank Saul Roseman and Norman D. Meadow of the Department of Biology of The Johns Hopkins University for supplying IIAGlc, Ernesto Freire of the Departments of Biology and Biophysics of The Johns Hopkins University for providing the results of calculations performed on the EcGK–IIAGlc complex, and Benjamin Lasseter and James Hu for helpful discussions. This study was supported by National Institutes of Health Grant GM-49992, Robert A. Welch Foundation Grant A-1479, and Texas Agricultural Experiment Station Grant H-6559. A.C.P. was supported in part by National Institutes of Health Chemistry/Biology Interface Training Grant T32-GM088523.
Abbreviations
FBP, fructose 1,6-bisphosphate
EcGK, E. coli glycerol kinase
HiGK, H. influenzae glycerol kinase
Chimeric enzyme nomenclature: The chimeric enzymes are named according to the portions of primary structure from each of the parent enzymes; for example, H:E245 is joined at amino acid 245 with the N-terminal portion from HiGK and the C-terminal portion from EcGK, and HII:E423–442 is constructed by replacing amino acids 423–442 in HiGKII with those from EcGK. The II denotes the amino acid residues identified crystallographically to interact with IIAGlc (7). Single-letter amino acid abbreviations following a hyphen after the name of the chimera, e.g., HiGK–GTR, show the identities of the residues at positions 427–429. Individual amino acid substitutions are denoted by using the single-letter abbreviations, e.g. G427D indicates replacement of the glycine at 427 by aspartate.
References
- 1.Pettigrew D. W. (1999) in Encyclopedia of Molecular Biology, ed. Creighton, T. (Wiley, New York), pp. 829–835.
- 2.Ito T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M. & Sakaki, Y. (2001) Proc. Natl. Acad. Sci. USA 98, 4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin E. C. C. (1996) in Escherichia coli and Salmonella: Cellular and Molecular Biology, ed. Neidhardt, F. C. (Am. Soc. Microbiol., Washington, DC), pp. 307–342.
- 4.Postma P. W., Lengeler, J. W. & Jacobson, G. R. (1993) Microbiol. Rev. 57, 543-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pettigrew D. W., Meadow, N. D., Roseman, S. & Remington, S. J. (1998) Biochemistry 37, 4875-4883. [DOI] [PubMed] [Google Scholar]
- 6.Feese M., Pettigrew, D. W., Meadow, N. D., Roseman, S. & Remington, S. J. (1994) Proc. Natl. Acad. Sci. USA 91, 3544-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hurley J. H., Faber, H. R., Worthylake, D., Meadow, N. D., Roseman, S., Pettigrew, D. W. & Remington, S. J. (1993) Science 259, 673-677. [PubMed] [Google Scholar]
- 8.Hurley J. H. (1996) Annu. Rev. Biophys. Biomol. Struct. 25, 137-162. [DOI] [PubMed] [Google Scholar]
- 9.Gerstein M., Lesk, A. M. & Chothia, C. (1994) Biochemistry 33, 6739-6749. [DOI] [PubMed] [Google Scholar]
- 10.Bystrom C. E., Pettigrew, D. W., Branchaud, B. P., O'Brien, P. & Remington, S. J. (1999) Biochemistry 38, 3508-3518. [DOI] [PubMed] [Google Scholar]
- 11.Feese M. D., Faber, H. R., Bystrom, C. E., Pettigrew, D. W. & Remington, S. J. (1998) Structure (London) 6, 1407-1418. [DOI] [PubMed] [Google Scholar]
- 12.Pawlyk A. C., (2001) Ph.D. thesis (Texas A&M University, College Station).
- 13.Lockless S. W. & Ranganathan, R. (1999) Science 286, 295-299. [DOI] [PubMed] [Google Scholar]
- 14.Komiyama N. H., Miyazaki, G., Tame, J. & Nagai, K. (1995) Nature (London) 373, 244-246. [DOI] [PubMed] [Google Scholar]
- 15.Guex N. & Peitsch, M. C. (1997) Electrophoresis 18, 2714-2723. [DOI] [PubMed] [Google Scholar]
- 16.Pawlyk A. C. & Pettigrew, D. W. (2001) Protein Expression Purif. 22, 52-59. [DOI] [PubMed] [Google Scholar]
- 17.Ormö M., Bystrom, C. E. & Remington, S. J. (1998) Biochemistry 37, 16565-16572. [DOI] [PubMed] [Google Scholar]
- 18.de Riel J. K. & Paulus, H. (1978) Biochemistry 17, 5141-5146. [DOI] [PubMed] [Google Scholar]
- 19.Liu W. Z., Faber, R., Feese, M., Remington, S. J. & Pettigrew, D. W. (1994) Biochemistry 33, 10120-10126. [DOI] [PubMed] [Google Scholar]
- 20.Pettigrew D. W., Liu, W. Z., Holmes, C., Meadow, N. D. & Roseman, S. (1996) J. Bacteriol. 178, 2846-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pettigrew D. W., Smith, G. B., Thomas, K. P. & Dodds, D. C. (1998) Arch. Biochem. Biophys. 349, 236-245. [DOI] [PubMed] [Google Scholar]
- 22.Hudson J. W., Golding, B. R. & Cerar, M. M. (1993) J. Mol. Biol. 234, 700-721. [DOI] [PubMed] [Google Scholar]
- 23.Rath V. L., Lin, K., Hwang, P. K. & Fletterick, R. J. (1996) Structure (London) 4, 463-473. [DOI] [PubMed] [Google Scholar]
- 24.Luque I. & Freire, E. (2000) Proteins 4,Suppl., 63-71. [DOI] [PubMed] [Google Scholar]
- 25.Higuchi R., Krummel, B. & Saiki, R. K. (1988) Nucleic Acids Res. 16, 7351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pettigrew D. W., Ma, D.-P., Conrad, C. A. & Johnson, J. R. (1988) J. Biol. Chem. 263, 135-139. [PubMed] [Google Scholar]
- 27.Holtman C. K., Pawlyk, A. C., Meadow, N. D. & Pettigrew, D. W. (2001) J. Bacteriol. 183, 3336-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Philo J. S. (1997) Biophys. J. 72, 435-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holtman C. K., Pawlyk, A. C., Meadow, N., Roseman, S. & Pettigrew, D. W. (2001) Biochemistry 40, 14302-14308. [DOI] [PubMed] [Google Scholar]
- 30.Lee B. & Richards, F. M. (1971) J. Mol. Biol. 55, 379-400. [DOI] [PubMed] [Google Scholar]
- 31.Hilser V. J. & Freire, E. (1996) J. Mol. Biol. 262, 756-774. [DOI] [PubMed] [Google Scholar]
- 32.Freire E. (2000) Proc. Natl. Acad. Sci. USA 97, 11680-11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Darbon E., Ito, K., Huang, H. S., Yoshimoto, T., Poncet, S. & Deutscher, J. (1999) Microbiology 145, 3205-3212. [DOI] [PubMed] [Google Scholar]
- 34.Lee N., Patrick, J. W. & Barnes, N. B. (1970) J. Biol. Chem. 245, 1357-1361. [PubMed] [Google Scholar]