Abstract
Sequence-specific binding of divalent cations to nucleosomal DNA can potentially influence nucleosome position and mobility, as well as modulate interactions with nuclear factors. We define the bonding and specificity of divalent cation interaction with nucleosomal DNA by characterizing Mn2+ binding in the x-ray structure of the nucleosome core particle at 1.9-Å resolution. Manganese ions are found ligated with high occupancy in the major groove to 12 of 22 GG and GC base pair steps. The specific location and mode of metal binding is the consequence of unambiguous conformational differences between dinucleotide sites, owing to their sequence context and orientation in the nucleosome core.
Divalent cations serve critical functions across a multitude of biochemical processes (1, 2). They generally play one of two roles: (i) bond activation in cleavage or rearrangement reactions of enzymes, or (ii) structural stabilization in macromolecules. The importance of divalent cations in the three-dimensional structure of DNA and RNA has been demonstrated in recent studies (3–6). A striking example is the induction of the B- to Z-form transition in poly[d(G-C)] by transition metal cations (7). In many crystal structures of nucleotides and oligonucleotides, divalent cations are observed to bind in a context or site-specific manner, interacting variably with base, phosphate, and sugar moieties depending on the nature of the metal ion. The recent crystal structures of two B-form DNA decamers at 1-Å resolution revealed up to 22 divalent cations per duplex, providing a detailed view of cation association to naked DNA and indicating that major groove binding of Mg2+ and Ca2+ can stabilize bending into the major groove (5).
The affinity of a cation for a specific site on a polynucleotide is a general function of its charge, hydration-free energy, coordination geometry, and coordinate bond-forming capacity (2, 8). The hydration-free energy is particularly important in distinguishing monovalent from divalent cation binding. Monovalent cations are generally less strongly solvated than divalent cations and therefore tend to interact with DNA purely electrostatically without making hydrogen bonds from metal-coordinated water molecules (5). As a consequence, DNA-associated monovalent cations are generally not clearly identified in x-ray crystallographic studies of duplex DNA (9). Although monovalent cations have been found in a few cases to be localized at preferred sites with low occupancies, the stable and specific solvation geometries adopted by divalent cations endow them with the ability to strongly and selectively bind DNA (10, 11).
Mg2+ is the most prevalent intracellular divalent cation, occurring in millimolar quantity (≈40 mM) (12). However, others, such as Mn2+ (≈0.5 μM) and Ca2+ (≈0.1 μM), have essential biological roles even though present at micromolar concentrations (13). These abundances in vivo in conjunction with observed effects on DNA conformation in vitro imply that divalent cations are of paramount importance for an accurate understanding of DNA structure and function. Structural studies investigating the role of these ions have to date focused on oligonucleotides alone. The DNA in eukaryotic cells is in contrast packaged in chromatin. The fundamental repeating unit of chromatin is the nucleosome, in which 147 bp of the variable repeat of roughly 200 bp are bound to the histone octamer in the nucleosome core (14). We have determined the 1.9-Å resolution structure of the nucleosome core particle (NCP), comprised of the four pairs of core histones (H3, H4, H2A, and H2B) and a 147-bp DNA (15). We present here the structural basis of how divalent cations interact with DNA in the nucleosome core based on the Mn2+ ions present in the crystals.
Methods
Crystal preparation, data collection, model refinement, and figure preparation have been described (15). The 147-bp NCP coordinates are available from the Protein Data Bank as file . Metal ions were initially located in Fo − Fc electron density maps as solvent peaks (>3σ) with the surrounding molecular environment accommodating one or more potential hydrogen bonds to the peak. Mn2+ ions were identified based on the residual peak (>3σ) occurring in Fo − Fc difference maps subsequent to full refinement with a water molecule at that position, and as significant peaks (>3σ) in anomalous difference maps. Additionally, water molecules refined at sites of metal ion binding attained B-factor values that were unreasonably low compared with the surrounding DNA atoms to which they were associated. The metal ions found were confirmed by final anomalous difference maps calculated by using phases from the fully refined model both with and without metal ions.
Assignment of DNA-bound metal ion type as Mn2+ as opposed to K+ is based empirically on coordination geometry and ligation distances. Mn2+ is most frequently observed to be hexa-coordinated with octahedral ligation geometry, whereas K+ has a strong preference to be octa-coordinated with variable, often irregular, ligation geometry (2, 16, 17). The metal binding sites identified in this study all have octahedral coordination geometry. Furthermore, the 0.5-Å increase in ionic radius from Mn2+ to K+ allows distinction between the two based on ligand-metal distances (16). The Mn2+ to ligand distances presented here range from 2.15 to 2.81 Å, which are much shorter than the minimal K+ to ligand distances of 2.9 Å observed for potassium binding to oxygen in oligonucleotide DNA (11). The mean Mn2+ to water-oxygen distance found here is 2.4 Å, in full agreement with the corresponding values from high-resolution crystal structures of enzymes containing Mn2+ binding sites (18–21).
Results
Divalent Cations in the NCP X-Ray Structure.
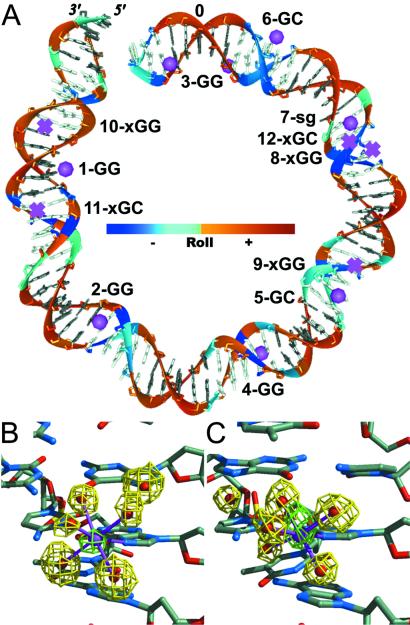
The structure determination of the NCP at 1.9-Å resolution has been described (15). Our crystals of the NCP were equilibrated in a solution containing 37 mM divalent cation as Mn2+ before x-ray data collection. The metal atoms and their coordinated water molecules were apparent in the 2Fo − Fc electron density map at early stages in the refinement. The Mn2+ ions were further identified by calculation of a difference Fourier map by using anomalous dispersion differences for structure factor amplitudes and phases corresponding to the refined atomic model lacking metal ions. DNA-bound Mn2+ ions were identified as map peaks with heights in the range of 3 to 9 times the rms difference from the mean density (σ) (Table 1, Fig. 1). Notably, the DNA sequence used here is 147 bp in length, increased from the 146-bp length used in our earlier studies at 2.8-Å and 2.0-Å resolution by addition of a single base pair inserted between the 2-fold symmetric halves (14, 15, 22). Although many of the divalent ions, including the Mn2+ ion bound to the H2A-H2B dimer, were seen in the 146-bp NCP at 2.0-Å resolution, the clarity of the electron density for the DNA and associated metal ions is substantially improved in the 147-bp structure.
Table 1.
Mn2+ sites in the nucleosome core particle
| Site | Location, bp | Sequence, 5′ to 3′ | Bond dist., Å | Nr. H2O ligands | Mean dist. to H2O, Å | Anom. signal, σ | Occupancy | B- factor, Å2 | Mode of binding | Roll, ° | Shift, Å | Slide, Å | Slide at 5′ TG, Å | 5′N7-3′O6 Dist., Å |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1-GG | 61 | AGGT | 2.6/2.5 N | 5/4 | 2.5/2.5 | 6.2/4.8 | 1.0/0.9 | 59/62 | 3-bond | 14.0 | −0.52 | −0.70 | — | 4.05 |
| 2-GG | 48 | TGGT | 2.4/2.5 N | 4/4 | 2.4/2.5 | 4.7/5.3 | 1.0/1.0 | 59/75 | 3-bond | 8.2 | −0.16 | −0.29 | 1.76 | 3.88 |
| 3-GG | −3 | TGGA | 2.5/2.4 N | 2/5 | 2.5/2.4 | 4.9/8.8 | 0.6/1.0 | 49/45 | 3-bond | 11.5 | −0.29 | −0.23 | 0.90 | 3.92 |
| 4-GG | −35 | TGGA | 2.4/2.6 N | 4/3 | 2.5/2.5 | 5.5/3.0 | 1.0/1.0 | 69/69 | 4-bond | 5.6 | 1.04 | 0.10 | 1.55 | 3.31 |
| 2.6/2.4 O | ||||||||||||||
| 5-GC | 27 | AGCA | 2.4/2.5 N | 5/3 | 2.3/2.6 | 7.6/6.2 | 1.0/0.7 | 52/64 | 3-bond | −5.6 | −1.01 | 0.15 | — | — |
| 6-GC | 5 | AGCT | 2.4/2.5 O | 4/3 | 2.5/2.3 | 3.2/4.4 | 0.7/0.6 | 67/45 | 4-bond | −10.1 | −1.18 | 0.20 | — | — |
| 7-sg | 13 | ATG | 6 | 2.3 | 6.7 | 0.7 | 35 | Minor grv. | ||||||
| 8-xGG | −16 | AGGC | Vacant | −10.8 | −0.96 | 0.71 | — | 4.40 | ||||||
| 9-xGG | 24 | TGGA | Vacant | −2.0 | 1.46 | −0.13 | 0.21 | 4.05 | ||||||
| 10-xGG | 64 | TGGA | Vacant | 6.2 | 0.48 | 0.12 | 0.17 | 4.02 | ||||||
| 11-xGC | 58\−59 | TGCA\TGCA | Vacant | 3.9 | −0.18\+0.18 | −0.47 | 0.72\1.63 | — | ||||||
| 12-xGC | 14\−15 | TGCC\GGCA | Vacant | −13.0 | −1.33\+1.33 | −0.08 | 0.18\— | — |
Unique Mn2+ binding sites. All sites are in the DNA major groove except for 7-sg in the minor (small) groove. GG and GC denote the type of base pair step binding the ion. x denotes a potential, but vacant Mn2+ site.
Distance in base pairs from the site of the central base pair (bp 0) on the molecular pseudo-dyad axis (15). Both chains run from base pair −73 at the 5′ terminus to +73 at the 3′ terminus. The 147-bp DNA is 2-fold sequence symmetric about bp 0. \ indicates the superposition of two GC sites.
Underlined nucleotide bases indicate the dinucleotide step defining Mn2+ binding sites in the major groove.
Bond distance from metal center to either the N7 or O6 liganded DNA atom. The two values correspond to the two halves of the DNA superhelix.
Parameter values are the average for two base pair steps, one from the identical sequence in each half of the DNA superhelix. DNA geometrical parameters are defined in ref. 39.
Fig 1.
Divalent cation binding sites on nucleosomal DNA. (A) Location of the Mn2+ binding sites in one half of the DNA superhelix (79 of 147 bp, 0 indicates the central base pair and 5′ and 3′ labels indicate the DNA terminus). All filled sites (•) occur at GG or GC base pair steps in the DNA major groove except for one site in the minor groove. The site numbers, including vacant GG and GC base pair steps (x), correspond to Table 1. DNA bases are shaded according to whether the major (gray) or minor (white) groove faces inward. The DNA backbone is colored according to the value of the base pair-step roll parameter as indicated (Roll). (B and C) Electron density for Mn2+ sites 1-GG and 5-GC, respectively. Anomalous difference electron density (green, 4σ contour) denotes the location of the Mn2+ ions, and simulated annealing, omit difference electron density (yellow, Fo − Fc 1.5σ and 2σ contours, respectively) corresponds to the positions of the metal-coordinated water molecules (five first shell, one second shell). Coordination bonds (magenta) between metal ion (magenta) and water molecules (red) are shown in the context of the surrounding base pair steps.
Sites of Mn2+ Binding.
The manganese ions bound in the NCP–DNA major groove interact exclusively with the N7 and O6 atoms of guanine at GG and GC steps either directly or via the water molecules in their first coordination shell. Octahedral coordination geometry is observed in all cases for the metal ions. The major groove edge of the guanine base provides a particularly efficacious binding site for cations as a consequence of the pronounced electronegative potential associated with the N7 and O6 atoms (23–25). Concerning the interaction between a metal ion and coordination shell water molecules, the positive charge of the ion is effectively propagated outward via the hydrogen atoms of the bound water molecules, which are oriented to interact with acceptor species. As we observe here in the NCP structure, the dense and extensive electronegative zone associated with GG and GC base pair steps provide favored sites for hydrogen bonding between the N7 and O6 acceptors and the strongly hydrated divalent cation. There are multiple occurrences in the NCP–DNA sequence for nine of the 10 different types of base pair steps: only the CG dinucleotide has zero occurrences. However as noted previously, CG steps are disfavored for cation binding because of the positions of the electropositive cytosine amino groups protruding into the major groove (26).
Modes of Binding in the Major Groove.
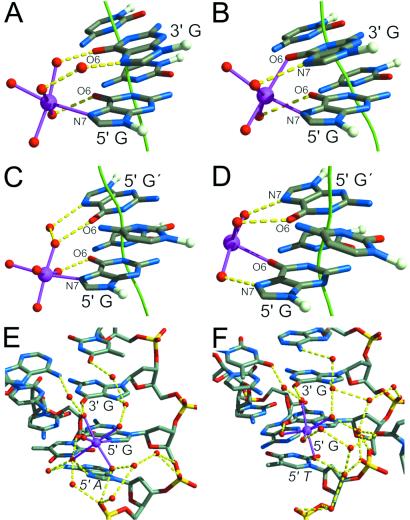
Two modes of Mn2+ binding occur in the DNA major groove, distinguished by either three or four DNA atoms that bind the metal ion-hydrate (Fig. 2 A–D). The three-bond and four-bond modes occur for both GG and GC base pair steps. In the case of the three-bond mode, one of the guanine bases (5′ for GG) is bound directly by its N7 atom to the metal ion, and indirectly by its O6 atom through a hydrogen bond from a metal-coordinated water molecule (Fig. 2 A and C). The O6 atom of the second guanine (3′ for GG) accepts a hydrogen bond from a second metal-coordinated water molecule. The N7 atom of this second guanine is linked to the same or a third metal-coordinated water molecule via a bridging water molecule. For four-bond GG, the coordination of the 5′ guanine is identical to the common arrangement in the three-bond mode. By comparison, the N7 and O6 atoms of four-bond GC have interchanged metal ion and metal-coordinated water molecule interactions (Fig. 2 B and D). The second guanine in four-bond GC has its N7 and O6 atoms each bound to a metal-coordinated water molecule, whereas for four-bond GG, the interaction between the O6 atom and metal is direct. The three-bond mode occurs six times for the GG base pair step and twice for the GC per complete NCP. There are two instances of the four-bond mode for both GG and GC steps.
Fig 2.
Three-bond and four-bond modes of Mn2+ binding in the DNA major groove. (A and C) The three-bond mode is shown for sites 1-GG and 5-GC, respectively. (B and D) The four-bond mode is shown for sites 4-GG and 6-GC, respectively. The guanine N7 and O6 atoms defining the metal binding mode and the contributing bases are labeled (G′ indicates the complementary base to C in the GC step). A nonzero shift parameter for a base pair step is visualized as a deviation of the double helix axis (green). Coordination bonds (magenta) between metal ion (magenta) and water molecules (red), and hydrogen bonds (yellow) between bases and first and second shell, coordinated water molecules are shown. (E) The site 1-GG (A) is shown with its full hydrogen bonding network. A hydrogen bond occurs between the N7 atom of the adenine base (5′ A) adjacent to the GG base step and a metal-coordinated water molecule, contributing to the orientation of the metal ion-hydrate. (F) Site 3-GG binds Mn2+ in the same three-bond mode as site 1-GG, but lacks the additional coordination shell bond from the adjacent thymine base (5′ T).
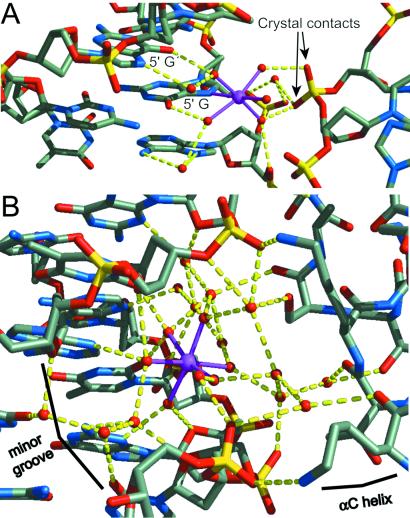
In addition to the local interactions occurring between metal, coordinate water ligands, and guanine bases, extensive hydrogen-bonding networks are observed in all cases between the metal-coordinated water molecules, further water molecules, neighboring bases, and phosphate groups (Figs. 2 E and F and 3). Significant differences in the orientation of the Mn2+-hydrate groups and their solvent-associated hydrogen-bonding pattern coincide with differences in the surrounding DNA sequence context and backbone conformation. Comparing three-bond sites 1-GG and 3-GG reveals the influence of the interaction between the N7 atom of the adjacent 5′ adenine and a metal-coordinated water molecule on the orientation of the Mn2+ octahedron (Fig. 2 E and F). Thus, DNA sequence context will affect cation binding, and in turn, cation binding may have some influence on DNA conformation. In another example, a Mn2+ ion bound at one of the two 5-GC sites directly mediates a crystal packing interaction, wherein two of the metal-coordinated water molecules serve as hydrogen bond donors to a phosphate group from a neighboring particle (Fig. 3A). The crystal contact is not requisite for Mn2+ binding, however, as the 5-GC site in the opposite half of the NCP also binds the cation without additional phosphate interaction. This type of bridging interaction may be important for compaction of nucleosomes into a higher-order structure and for stabilization of the chromatin fiber. The general dependency of chromatin compaction on ion type and concentration has been described (27–29).
Fig 3.
Metal ions bound between particles. (A) The Mn2+ ion in site 5-GC makes a crosslink to an adjacent NCP in the crystal. Additional interactions occur between the metal-coordinated water molecules and phosphate oxygen atoms. (B) Mn2+ binding in the DNA minor groove. The metal ion is fully hexahydrated and therefore makes no direct bonds with DNA. Three metal-coordinated water molecules interact with two thymine O2 atoms, an adenine and a guanine N3 atom, and two deoxyribose O4′ atoms. The remaining three water molecules are linked to further water molecules bound to a neighboring NCP. The divalent cation partially mediates the crystal contact between the minor groove of one particle and a H2B αC-helix of another.
Single Site in the Minor Groove.
A Mn2+ ion binds in the minor groove at only one location in the entire NCP structure. In contrast to those bound in the major groove, the minor groove-bound ion maintains its full octahedral hydration shell. This hexahydrate cation is bound between the surface of the minor groove and the H2B αC-helix coming from a neighboring NCP in the crystal (Fig. 3B). The three water ligands that are positioned deepest into the minor groove form an extensive network of hydrogen bonds with the acceptor groups of adenine, thymine, and guanine as well as deoxyribose O4′ atoms. The interactions of the other three metal-coordinated water molecules with the histone are mediated by intervening solvent layers. The symmetric site in the other half of the nucleosomal DNA is not juxtaposed with another NCP and does not have a Mn2+ cation bound. Internucleosomal histone–cation–DNA interactions, as observed here in our crystals and as suggested from sedimentation analysis of oligonucleosomes (30), may mediate chromatin compaction in addition to DNA–cation–DNA interactions. Internuclesomal histone–cation–histone interactions also occur in the crystals (15).
Mn2+ Binding Mode Selectivity.
Both GG and GC base pair steps bind Mn2+ in the three-bond and four-bond modes. The mode of binding occurring at a particular site does not appear to depend on further solvent interactions with adjoining base pairs. The selectivity is a consequence of the precise conformation of the base pairs in the site arising from histone-imposed constraints and sequence contextual effects. The conformational response of each base pair step accommodating DNA superhelix formation in the NCP depends both on the precise histone interactions and the local DNA sequence (unpublished work).
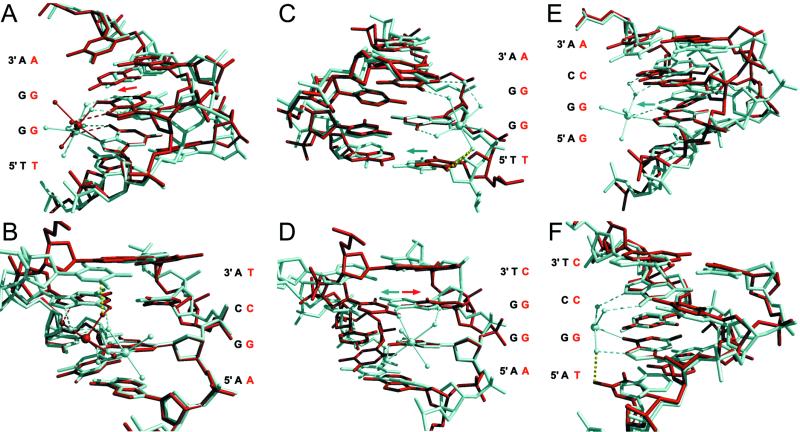
The sites 1-, 2-, and 3-GG have Mn2+ bound in the three-bond mode whereas site 4-GG binds the cation in the four-bond mode. The base pair-step shift parameter facilitates the four-bond mode in this case. The GG base pair step of site 4-GG is strongly positively shifted, and as a consequence, the 3′ guanine protrudes further into the major groove than the adjacent 5′ guanine (Fig. 4A). This arrangement permits entry of the O6 atom of the 3′ guanine into the metal coordination shell whereas the direct metal interaction with the N7 atom of the 5′ guanine is maintained. Both of these sites are contained in the sequence TGGA, demonstrating the importance of the conformation imposed by the nucleosome core on divalent cation binding.
Fig 4.
Comparisons of Mn2+-binding sites. Alignments of pairs of metal-binding sites show that DNA base pair-step conformational differences govern metal binding. Selected guanine bases (11 atoms) from two sites are aligned by least-squares superposition. The metal ion and water molecules (spheres) are shown with their coordinate bonds (solid) and hydrogen bonds (dashed). The sequence of the DNA strands having the direct metal bond(s) is indicated. (A) The 3-GG (white) and 4-GG (red) occupied sites demonstrate the preference for either three-bond or four-bond mode, respectively, for GG base pair steps. The GG base pair step for site 4-GG is strongly positively shifted (red arrow) as compared with that for 3-GG. (B) The 5-GC (white) and 6-GC (red) occupied sites illustrate the origin of three-bond versus four-bond modes for GC base pair steps. The virtual steric clash (yellow dotted line) between a metal-coordinated water molecule at site 6-GC (four-bond) and the C5 atom of the thymine base flanking site 5-GC (three-bond) is shown. (C) The occupied 3-GG (white) and vacant 10-xGG (red) sites show the importance of a 5′ flanking thymine for GG steps. Substantial positive slide for the TG step (white arrow) in the TGG sequence is required to avoid steric interference (yellow dotted line) between a metal-coordinated water molecule and the C5-methyl and C6 groups of the flanking thymine. Pronounced positive slide occurs at TGG sites 2-GG, 3-GG and 4-GG, but not at 9-xGG and 10-xGG. (D) The 1-GG (white) and 8-xGG (red) sites demonstrate the effect of roll-slide anticorrelation. Positive values for roll occurring for bending into the major groove result in low slide values (white arrow) and favor metal binding. Bending into the minor groove results in sliding in the opposite direction (red arrow) and disfavors metal binding by reducing stacking of the guanine bases. (E) The 5-GC (white) and 12-xGC (red) sites illustrate the influence of the shift parameter on GC sites. The guanine base in the sequence-symmetric GC base pair step must have a large negative shift value (white arrow) for it to be bound directly to a metal ion. This shift value causes it to protrude into the major groove relative to its counterpart on the opposite strand. (F) The 6-GC (white) and 12-xGC (red) sites show the importance of a 5′ flanking thymine for GC steps. For metal binding to occur to the guanine in the sequence TGC, the TG step must have sufficient positive slide to avoid the steric clash (yellow dotted line) between the C5-methyl group of the thymine and a metal-coordinated water molecule.
For the GG base pair step, the N7 atom of 5′ guanine always makes a direct interaction with the metal ion. In the four-bond scheme, the 5′ guanine N7 and 3′ guanine O6 atoms are the only pair within distance (3.31 Å for site 4-GG, Table 1) to permit the metal-coordinated water molecules to bind the remaining O6 and N7 atoms. For three-bond GG, the distance (4.05 Å for site 1-GG) between 5′ guanine N7 and 3′ guanine O6 atoms increases, preventing two direct metal-base bonds. The bonding solution adopted is localization of a metal-coordinated water molecule between the 3′ guanine O6 and metal atoms. Either the higher affinity of the nitrogen over the oxygen atom for the Mn2+ ion or more subtle bond geometry optimization results in the favored metal-N7 atom bond.
One example of three- and four-bond mode for GC base pair steps occurs in each half of the DNA superhelix at sites 5-GC and 6-GC, respectively. The predominant difference between these sites lies in the thymine complementary to the 3′ adenine in the 5-GC site versus adenine for the 6-GC site. This flanking thymine would sterically interfere with one of the metal-coordinated water molecules if it were bound in four-bond mode (Fig. 4B). Therefore, the DNA sequence and its conformation impose three-bond mode on the metal ion in site 5-GC. Because of the additional bond with the metal-hydrate, the four-bond mode seen for site 6-GC may be the preferred mode of binding at GC steps lacking steric hindrance from a 3′ AT base pair, however.
Occupied Versus Vacant Sites.
Of the 11 potential GG and GC dinucleotides that occur in each half of the NCP DNA sequence, only six are occupied by a Mn2+ cation. The remainder are vacant or have occupancy too low to be observed. Here, as in the selection of binding mode, histone- and sequence-dependent conformational variation appears to be responsible for the differences. For example, of the five unique sites containing the trinucleotide TGG, three are occupied and two are vacant, and it is the slide parameter of the TG base pair step preceding the metal binding site that is responsible. If the value of slide for the TG step is 0.9 Å or larger, then Mn2+ is bound, and when less, the site is vacant (Table 1). The actual slide value depends on the location of the base pair step in the NCP superhelix (unpublished work). As shown by superposition of the structures of sites 3-GG and 10-xGG, the positive slide value circumvents a steric clash between a metal-coordinated water molecule and the C5-methyl and C6 groups of the thymine base (Fig. 4C). In contrast, site 1-GG has the sequence AGG, and the additional bond between the metal-coordinated water molecule and adenine N7 atom, as discussed, stabilizes the ion at this site (Fig. 2E).
More generally, Mn2+ binding to GG steps is correlated with positive roll, that is, the ion does not bind the major groove when it is facing directly away from the histone octamer. All occupied GG sites have a positive value for the base pair-step roll angle (Table 1). Alignment of sites 1-GG and 8-xGG shows how the orientation of the bases in the step influences metal binding (Fig. 4D). For both GG and GC base steps, roll-slide conformational coupling can improve the alignment of the electronegative zones of the guanine bases and their liganding atoms. This is particularly noticeable for GC steps with bound metal that, in contrast to GG steps, have the major groove facing away from the histone octamer and have a negative roll value (Table 1). The corresponding positive slide value brings the guanine bases on opposing DNA strands in the GC step closer together.
Of the four distinct GC sites, 5-GC and 6-GC contain a manganese ion and 11-xGC, and 12-xGC do not. In addition, each GC dinucleotide can be viewed as two overlapping sites read in opposite directions, with metal binding to one site, defined by the 5′ guanine making a direct metal interaction, excluding metal binding to the other. The same principles that govern occupation versus vacancy may also apply to this selection. Both sites 5-GC and 6-GC display a large negative shift value, and as a consequence the guanine base of the step that protrudes further into the major groove contributes its N7 or O6 atom directly to the metal coordination shell (Fig. 2C and D, Table 1). The superposition of the 5-GC and 12-xGC sites demonstrates this effect (Fig. 4E). The 12-xGC site read in the direction GGCA, corresponding to the AGCA for the 5-GC site, has a large positive shift which is opposite to the direction that favors metal binding.
Alignment of the 12-xGC sequence read in the TGCC direction with site 6-GC reveals that the preceding TG step is effective in blocking metal binding (Fig. 4F). The C5-methyl group would sterically interfere with a metal-coordinated water molecule. This conformational effect can also be seen by superposition of the 5-GC site with itself in the opposite orientation (not shown). Unlike the TGG sequences allowing metal binding because of their large positive slide at the TG step, the value of slide is near zero for 12-xGC. For the 11-xGC site, the GC step is flanked by TG steps in both directions. Although both TG steps have high positive slide, this site is vacant owing to its near zero shift value and negative slide value.
Discussion
Divalent Cations and DNA Binding.
Our observations on divalent cation binding in the NCP are consistent with previous studies on naked DNA. Investigation of Mn2+, Zn2+, Co2+, and Ni2+ interaction with oligonucleotide DNA by NMR indicated that the strongest preference in binding is for the 5′ guanine N7 atom of GG steps (31, 32). Likewise, crystal structures have shown that Mg2+ and Ca2+ bind to the major groove predominantly at GG steps (5). The pronounced affinity of divalent cations for the GG dinucleotide is readily understandable on purely electrostatic grounds, as was indicated earlier by theoretical calculations (23). However, the precise mode of metal coordination is largely determined by where the metal ion lies on the bonding Pearson hardness/softness scale, and although Mg2+, Ca2+, and Mn2+ all are regarded as hard ions, the relative degree varies significantly (33). These differences explain why the Ca2+ ion is observed to ligate directly to the relatively soft N7 atom of guanine, whereas the more difficult to dehydrate, harder Mg2+ ion interacts only with such moieties via water molecules (5, 34). The Mn2+ ion, on the other hand, is softer than Ca2+ and has a higher affinity for the guanine N7 atom, consistent with our findings for nucleosomal DNA (7, 35). Interestingly, the observed mode of Ca2+ ligation to the major groove side of GG steps (5, 34) in oligonucleotide structures is virtually identical to that observed for Mn2+ binding to site 4-GG in the NCP, in which the metal ion is ligated directly to both the 5′ guanine N7 and 3′ guanine O6 atoms. In this case, the relative softness of the ions and their dehydration energies appear to be more important in determining their similarity in binding than their ligation geometry (36). The Mg2+ ion, like Mn2+, is hexa-coordinate, whereas Ca2+ is hepta-coordinate.
Conformation Dictates the Ion Binding Mode.
Mn2+ ion binding in the DNA major groove occurs exclusively at GG and GC base pair steps in the NCP structure; however, selection of the subset of these sites that are occupied is governed by the local DNA conformation. The metal ion itself has not completely, if at all, overridden the DNA conformation resulting from the sequence context and histone interactions as six of 14 GG steps and four of eight GC steps do not have divalent ion bound. Indeed, the conformation of the base pairs adjacent to the metal-binding step can exclude ion binding. Even when a site is occupied, the metal ion adjusts its coordination shell to suit the precise geometry of the GG or GC base pair step and flanking base pairs so that one or two water molecules are replaced by heterocyclic atoms of the guanine bases. Owing to observed conformational similarities for base pair steps dependent on their location and thus their orientation in the NCP, DNA conformation appears to dictate metal binding, as opposed to the converse (unpublished work). These influences of orientation, which are unique features of the systematically wrapped DNA in chromatin, can determine the capacity for metal ion binding.
Although metal ion binding is evidently unable to dominate over the intrinsic sequence-dependent conformation of NCP DNA, bound metal ion generally will contribute to the stabilization of the preexisting conformation. The strong anticorrelation of the base pair-step roll and slide parameters noted in structures of DNA oligonucleotides is also apparent for the NCP DNA (37) (unpublished work). This inherent feature of DNA conformation can increase the overlap of negative electrostatic potential emanating from the guanine base edges in the major groove. The metal-bound GG steps are rolled into the major groove and show low slide values whereas the metal-bound GC steps are rolled into the minor groove and show high slide values. The resulting localized, higher charge density favors ion binding in the major groove in either case, and conversely ion binding will stabilize these conformations. The stabilization of bending into the major groove seen in oligonucleotides at GG steps has been noted for Mg2+ and Ca2+ ions (5). The effect is apparently much weaker at GC steps in oligonucleotides versus the NCP, however, reflecting the general preference for major groove bending in oligonucleotides compared with the NCP (31).
Conclusion
The contribution of divalent ion binding to the stability of the nucleosome and its selection of positions along genomic DNA sequences requires further investigation, but the structural data presented here suggest a significant role based on the placement and context of GG and GC base pair steps in the DNA sequence. High-resolution mapping of nucleosome position (38) in solutions containing differing concentrations and types of metal ions would be illuminating. Equally important, special sites that occur in the DNA sequence of a particular nucleosome, such as the 5-GC and 7-sg sites seen here, may confer internucleosomal interactions that locally influence the stability of the higher-order structure of nucleosomes in the chromatin fiber.
Acknowledgments
We are grateful to Dr. Imre Berger and Dr. David Sargent for their comments on the manuscript and appreciate the support of the Swiss National Fund.
Abbreviations
NCP, nucleosome core particle
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Holm R. H., Kennepohl, P. & Solomon, E. I. (1996) Chem. Rev. 96, 2239-2314. [DOI] [PubMed] [Google Scholar]
- 2.Glusker J. P. (1991) Adv. Protein Chem. 42, 1-76. [DOI] [PubMed] [Google Scholar]
- 3.Cate J. H., Hanna, R. L. & Doudna, J. A. (1997) Nat. Struct. Biol. 4, 553-558. [DOI] [PubMed] [Google Scholar]
- 4.Minasov G., Tereshko, V. & Egli, M. (1999) J. Mol. Biol. 291, 83-99. [DOI] [PubMed] [Google Scholar]
- 5.Chiu T. K. & Dickerson, R. E. (2000) J. Mol. Biol. 301, 915-945. [DOI] [PubMed] [Google Scholar]
- 6.Sines C. C., McFail-Isom, L., Howerton, S. B., VanDerveer, D. & Williams, L. D. (2000) J. Am. Chem. Soc. 122, 11048-11056. [Google Scholar]
- 7.van de Sande J. H., McIntosh, L. P. & Jovin, T. M. (1982) EMBO J. 1, 777-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Misra V. K. & Draper, D. E. (1998) Biopolymers 48, 113-135. [DOI] [PubMed] [Google Scholar]
- 9.Chiu T. K., Kaczor-Grzeskowiak, M. & Dickerson, R. E. (1999) J. Mol. Biol. 292, 589-608. [DOI] [PubMed] [Google Scholar]
- 10.Howerton S. B., Sines, C. C., VanDerveer, D. & Williams, L. D. (2001) Biochemistry 40, 10023-10031. [DOI] [PubMed] [Google Scholar]
- 11.Tereshko V., Wilds, C. J., Minasov, G., Prakash, T. P., Maier, M. A., Howard, A., Wawrzak, Z., Manoharan, M. & Egli, M. (2001) Nucleic Acids Res. 29, 1208-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins K. D. (1997) Biophys. J. 72, 65-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christianson D. W. (1997) Prog. Biophys. Mol. Biol. 67, 217-252. [DOI] [PubMed] [Google Scholar]
- 14.Luger K., Maeder, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. (1997) Nature (London) 389, 251-260. [DOI] [PubMed] [Google Scholar]
- 15.Davey C. A., Sargent, D. F., Luger, K., Maeder, A. W. & Richmond, T. J. (2002) J. Mol. Biol. 319, 1097-1113. [DOI] [PubMed] [Google Scholar]
- 16.Brown I. D. (1988) Acta Crystallogr. B 44, 545-553. [Google Scholar]
- 17.Harding M. M. (1999) Acta Crystallogr. D 55, 1432-1443. [DOI] [PubMed] [Google Scholar]
- 18.Sundaramoorthy M., Kishi, K., Gold, M. H. & Poulos, T. L. (1994) J. Biol. Chem. 269, 32759-32767. [PubMed] [Google Scholar]
- 19.Kanyo Z. F., Scolnick, L. R., Ash, D. E. & Christianson, D. W. (1996) Nature (London) 383, 554-557. [DOI] [PubMed] [Google Scholar]
- 20.Unligil U. M., Zhou, S., Yuwaraj, S., Sarkar, M., Schachter, H. & Rini, J. M. (2000) EMBO J. 19, 5269-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagner T., Shumilin, I. A., Bauerle, R. & Kretsinger, R. H. (2000) J. Mol. Biol. 301, 389-399. [DOI] [PubMed] [Google Scholar]
- 22.Luger K., Maeder, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. (2000) in Biomolecular Stereodynamics, ed. Sarma, R. H. (Adenine, New York), Vol. 11, pp. 190–200. [Google Scholar]
- 23.Pullman A. & Pullman, B. (1981) Q. Rev. Biophys. 14, 289-380. [DOI] [PubMed] [Google Scholar]
- 24.Hunter C. A. (1993) J. Mol. Biol. 230, 1025-1054. [DOI] [PubMed] [Google Scholar]
- 25.Packer M. J., Dauncey, M. P. & Hunter, C. A. (2000) J. Mol. Biol. 295, 71-83. [DOI] [PubMed] [Google Scholar]
- 26.Auffinger P. & Westhof, E. (2000) J. Mol. Biol. 300, 1113-1131. [DOI] [PubMed] [Google Scholar]
- 27.Widom J. (1986) J. Mol. Biol. 190, 411-424. [DOI] [PubMed] [Google Scholar]
- 28.Clark D. J. & Kimura, T. (1990) J. Mol. Biol. 211, 883-896. [DOI] [PubMed] [Google Scholar]
- 29.Blank T. A. & Becker, P. B. (1995) J. Mol. Biol. 252, 305-313. [DOI] [PubMed] [Google Scholar]
- 30.Schwarz P. M., Felthauser, A., Fletcher, T. M. & Hansen, J. C. (1996) Biochemistry 35, 4009-4015. [DOI] [PubMed] [Google Scholar]
- 31.Froystein N. A., Davis, J. T., Reid, B. R. & Sletten, E. (1993) Acta Chem. Scand. 47, 649-657. [DOI] [PubMed] [Google Scholar]
- 32.Moldrheim E., Anderson, B., Frøystein, N. A. & Sletten, E. (1998) Inorg. Chim. Acta 273, 41-46. [Google Scholar]
- 33.Pearson R. G. (1990) Coord. Chem. Rev. 100, 403-425. [Google Scholar]
- 34.Kielkopf C. L., Ding, S., Kuhn, P. & Rees, D. C. (2000) J. Mol. Biol. 296, 787-801. [DOI] [PubMed] [Google Scholar]
- 35.Zacharias W., Larson, J. E., Klysik, J., Stirdivant, S. M. & Wells, R. D. (1982) J. Biol. Chem. 257, 2775-2782. [PubMed] [Google Scholar]
- 36.Draper D. E. & Misra, V. K. (1998) Nat. Struct. Biol. 5, 927-930. [DOI] [PubMed] [Google Scholar]
- 37.El Hassan M. A. & Calladine, C. R. (1997) Philos. Trans. R. Soc. London A 355, 43-100. [Google Scholar]
- 38.Flaus A., Luger, K., Tan, S. & Richmond, T. J. (1996) Proc. Natl. Acad. Sci. USA 93, 1370-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dickerson R. E., Bansal, M., Calladine, C. R., Diekmann, S., Hunter, W. N. & Kennard, O. (1989) EMBO J. 8, 1-4.2714249 [Google Scholar]