Abstract
Understanding the gene-expression patterns during liver regeneration may help to reveal how regenerative processes are initiated and controlled as well as shed new light onto processes that lead to liver disease. Using high-density oligonucleotide arrays, we have examined the gene-expression program in the livers of mice after partial hepatectomy. A time course was constructed for gene expression between 0 and 4 h after partial hepatectomy, corresponding to the priming phase of liver regeneration. The genomic program for liver regeneration involves transcription-factor generation, stress and inflammatory responses, cytoskeletal and extracellular matrix modification, and regulation of cell-cycle entry. The genome-wide changes that are observed provide a detailed and comprehensive map of the initial priming stage of liver regeneration.
The liver's ability to regenerate in mammals is relatively unique. Only a few species, including certain worms, insects, reptiles, and amphibians, can readily undergo various types of reparative regeneration including epimorphic reconstruction of entire body parts. In contrast, humans and larger mammals have little regenerative capacity (1, 2). Examples of organs and body parts that show reasonably good regenerative capacities are few and include the liver, fingertips, and peripheral nerves, and stem cells may be a source of the regenerative capacity (2). Among these types of reparative processes, liver regeneration stands out. The liver is capable of modulating its mass according to functional requirements, proliferating under conditions of functional deficiency, and undergoing apoptosis under functional excess. In both of these processes, the liver undergoes remodeling of the entire organ physiology to preserve normal histological organization (3, 4). Liver regeneration does not rely on stem cells, although liver stem cells may contribute to the process, and each cell type has the capacity to enter into a proliferative state and also alter their differentiation so that liver cells have innate progenitor cell characteristics (5, 6). Hepatocytes are the first cell types to enter into the cell cycle after functional deficiency in the liver (4), and they show an almost limitless capacity to proliferate (7). Also, during liver regeneration the liver cells continue to perform crucial metabolic functions such as glucose regulation, synthesis of many blood proteins, the secretion of bile, and biodegradation of toxic compounds required for homeostasis (3). Understanding the molecular mechanisms and genomic program of liver regeneration is of fundamental importance and is the first step toward controlling these events and other regenerative processes.
Liver regeneration can be initiated in several ways. Classical methods for initiating liver regeneration in animal models involve either partial hepatectomy (PHx) or injection of hepatotoxic compounds such as CCl4. Pioneering studies by Taub and coworkers (8) as well as Fausto (3) have defined the roles of many immediate-early genes and cytokines in liver regeneration. On a molecular level, the entry of hepatocytes into cell cycle is stimulated by various cytokines and growth factors. Examples include IL-6, hepatocyte growth factor (HGF), epidermal growth factor, tumor necrosis factor (TNF)-α, transforming growth factor-α, insulin, and other receptor ligands that have been implicated in various stages of hepatocyte proliferation (3, 4). These ligands, through complex molecular mechanisms, activate transcription factors including NF-κB, signal transducer and activator of transcription 3 (STAT3), activator protein 1 (AP-1), and CCAAT/enhancer-binding protein (C/EBP)β that initiate a cascade of gene expression that ultimately is responsible for proliferation (9).
Before cell-cycle entry, quiescent hepatocytes (G0) undergo a priming phase (G0 → G1) during which the cells reenter the cell cycle and prepare for proliferation. The concept of priming phase in hepatocytes, originally proposed by Fausto et al. (3, 10–12), is the first stage of liver regeneration, the duration of which is species-dependent (13). For mice this stage lasts for ≈4 h after PHx. During this time, immediate-early genes such as c-fos and c-jun are induced (3, 8, 14, 15). In fact, many genes have been identified as being differentially expressed during hepatocyte priming and the following stages of the cell cycle leading up to DNA replication and mitosis. Known exogenous priming stimuli include sham surgery, protein deprivation, and collagenase treatment as well as infusion of TNFα, epidermal growth factor, or HGF (3, 4).
Given that the cells in a regenerating liver have progenitor cell character, we used functional genomic technologies to study cellular priming in mice. We have characterized the genome-wide expression changes in mice after ≈70% PHx over the course of 4 h, which corresponds to the priming stage of hepatocyte proliferation. Sham surgeries were conducted to eliminate responses that are caused by the surgery alone, and resected liver specimens functioned as a baseline for gene-expression changes in each mouse. We found that genes associated with transcription-factor production, stress and inflammatory responses, cytoskeletal and extracellular matrix modification, and regulation of cell-cycle entry all are involved in the early stages of liver regeneration. It is noteworthy that the liver is composed of many different types of cells, and most of its function is confined to hepatocytes (which represent ≈70% of the liver), Kupffer cells (macrophages), and bile ductule epithelium. Thus, it is possible that some of the changes reported may have occurred in nonparenchymal cells of the liver rather than hepatocytes.
Materials and Methods
Mice, Tissues, and RNA Preparation.
Groups of three 8–10-week-old male C57BL/6 mice were anesthetized and subjected to either sham operation or 70% PHx as described (16). At 1, 5, 10, 30, 90, and 240 min posthepatectomy, mice were killed, and liver tissue samples were harvested. Total RNA was isolated from these samples and the respective surgically resected liver sections as described (16) and analyzed by high-density oligonucleotide microarray, Northern blot, and RNase protection assay (RPA).
High-Density Oligonucleotide Microarray Analysis.
Double-stranded DNA was synthesized from ≈5 μg of total RNA by using the SuperScript Choice system (Life Technologies, Grand Island, NY) and a primer containing poly(dT) and a T7 RNA polymerase promoter sequence (Genset, La Jolla, CA). In vitro transcription using double-stranded cDNA as a template in the presence of biotinylated UTP and CTP was carried out by using an Ambion (Austin, TX) in vitro transcription kit. Biotin-labeled cRNA was purified, fragmented, and hybridized to Mu6.5K arrays (Affymetrix, Santa Clara, CA). The arrays were washed and stained with streptavidin-phycoerythrin and then scanned with an Affymetrix GeneChip scanner. Primary image analysis was performed by using the GENECHIP 3.1 software, and images were scaled to an average difference value of 200 as described previously. Hybridizations were performed in duplicate, and only differential expression observed in both replicates was analyzed further. Comparison analyses for data at each time point were calculated by using the t0 and sham as baselines. Gene-expression profiles were established from RNA samples isolated from four different mice per time point (two PHx and two sham).
Reverse Transcription–PCR, Northern Blots, and RPA.
Probes for Northern blotting were amplified from RNA in the 240-min PHx time point by using the following primer pairs: Pip92Fwd (5′-GAGTCTGCAGCTATCCCTCG-3′), Pip92Rev (5′-CACGTTGAGCATATTGTCGG-3′); Tis21Fwd (5′-CCTAGCCAAGGTAAAAGGGG-3′), Tis21Rev (5′-GGTCCTCTCCATCTTAGCC-3′); and glvr-1Fwd (5′-GGTGGGATGTGCAGTTTTCT-3′), glvr-1Rev (5′-CCTTGTGCACGGTGTGATAC-3′). RNA samples were analyzed by electrophoresis and transferred to nitrocellulose membranes. The 32P-labeled cDNA probes along with a 32P-labeled probe for the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) then were used to probe message levels by using established protocols (16). Quantitation of chemokines MCP-1 and GRO as well as L32 was performed also by RPA exactly as described previously (17, 18).
Results and Discussion
We have examined the gene-expression changes in the liver of mice after PHx over a time course spanning 4 h by using high-density oligonucleotide arrays with probes for ≈6,500 genes. Liver RNA samples were removed at 1, 5, 10, 30, 90, and 240 min after PHx. No reproducible effects were observed at 1 and 5 min after PHx (data not shown). The experiments showed that 185 genes had altered gene expression for at least one of the remaining time points (Table 1). All of these genes showed higher induction or repression in regenerating livers than in sham controls, indicating the importance of including these controls in our analysis. We have excluded genes that were induced or repressed at similar levels in sham and regenerating livers, because these gene-expression changes are likely indicative of the effects of the surgery. It is important also to note that a wide variety of the genes observed to be differentially regulated according to microarray analysis have not been described previously in more limited studies of hepatocyte priming during liver regeneration after PHx.
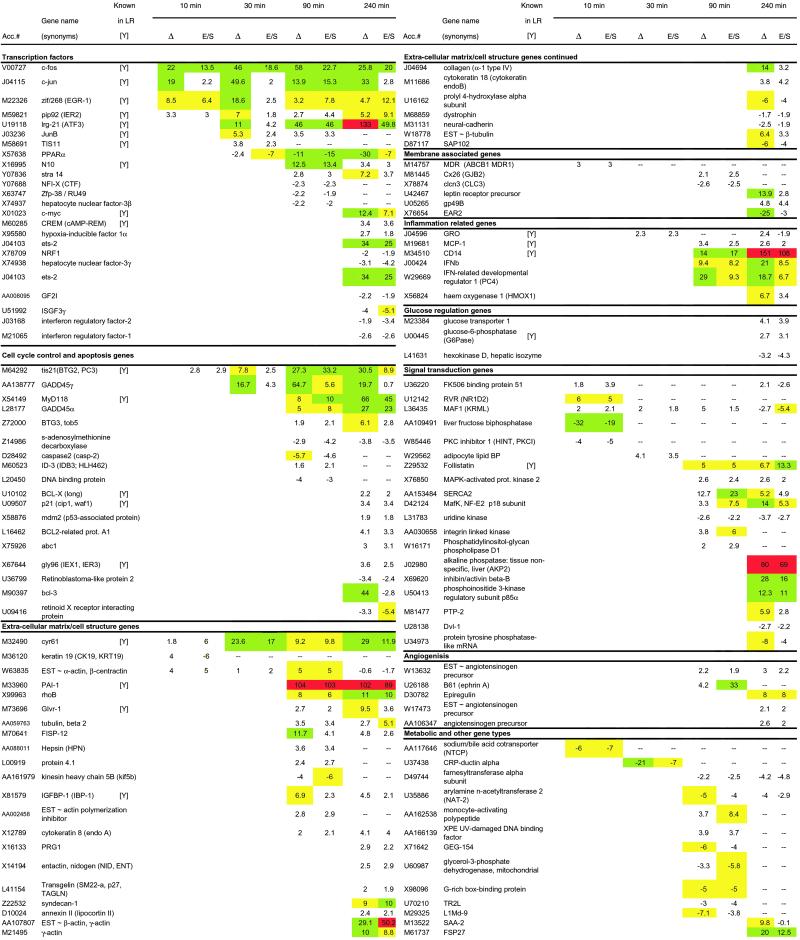
Table 1.
GeneChip analysis of changes in liver gene expression after partial hepatectomy
Fold change vs. resected liver specimens (Δ) for differentially expressed genes as well as the fold change vs. sham surgery (E/S) are listed in rows for time points 10, 30, 90, and 240 min. Genes are listed by GenBank accession number (Acc.#) and name and are grouped according to their function. The fold change for gene expression with values greater than 5 are highlighted; yellow corresponds to values >5; green corresponds to values >10; red corresponds to values >50. Genes that previously had been identified in liver regeneration, particularly in the priming stage prior to entry into cell cycle, are indicated with a [Y].
Transcription Factors.
During liver regeneration, liver cells are exposed to stresses associated with functional deficiency, and these stresses ultimately lead to cell proliferation (9, 19, 20). We have identified 19 immediate-early transcription-factor genes that are differentially regulated during the priming phase (Table 1), many of which overlap with previously established immediate-early genes implicated by Taub and coworkers (8, 15) and Fausto and coworkers (3, 10–12) that have established the importance of inflammation and protooncogenes in the early stages of liver regeneration. It is well established that liver regeneration involves the posttranslational activation of the transcription factors NF-κB, STAT3, AP-1, and C/EBPβ by mechanisms connected to increased levels of cytokines and reactive oxygen species within the liver (9, 15, 19). Immediate-early genes such as the archetypal immediate-early genes c-fos, c-jun, and c-myc are up-regulated as a consequence (9). Immediate-early transcription factors generated in response to PHx represent a critical step in controlling the proliferation of hepatocytes within a regenerating liver. Cooperatively, they activate the proliferative program within quiescent hepatocytes.
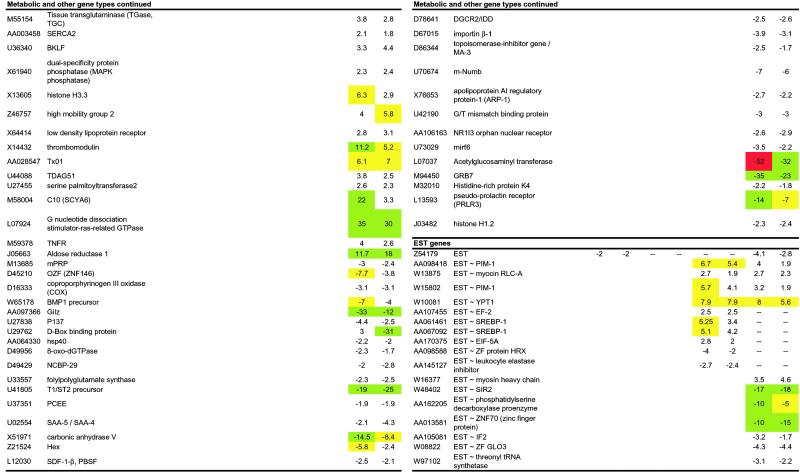
Four immediate-early transcription factors were activated as early as 10 min after PHx: c-fos, c-jun, Zif/268, and pip92. Of these, c-fos and c-jun are primary immediate-early genes expressed by many cell types in response to cellular stress. Zif/268 (EGR-1) contains three zinc fingers recognizing a GC-rich sequence, which has been identified in the promoter regions of a number of genes including PRL-1, a mitogenic phosphatase associated with regenerating livers that plays an important role in cell growth in a number of different tissue types (21). The other immediate-early transcription factor in the 10-min time point is PIP92, the induction of which has been confirmed by Northern blot (Fig. 1). PIP92 encodes a short-lived proline-rich protein with no sequence homology to other known proteins. PIP92 is known to be stimulated by cytokines such as fibroblast growth factor and by mitogen-activated protein kinase (MAPK) signaling pathways and has been implicated in processes such as differentiation and stimulation of fibroblasts (22).
Fig 1.
Induction of pip92, gro/KC, tis21, glvr-1, MCP-1, and C10 during the priming phase of liver regeneration. Age-matched C57BL/6 male mice (three mice per group) were subjected to 70% PHx and killed at different points afterward. Total RNA (20 or 10 mg) extracted from their livers were analyzed by Northern blot (NB) and RPA analyses, respectively, for the expression of pip92, gro/KC, tis21, glvr-1, MCP-1, and C10. Results from a representative mouse per group were compared with those obtained either in its own resected liver section (t0) or in mice that were subjected to sham operation and killed at the same time point. The housekeeping genes glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and ribosomal protein light 32 (L32) were used to normalize either the amount of RNA bound to the membrane (NB) or the amount of RNA loaded in each lane (RPA).
We observed high induction of ATF3, also known as liver-regenerating factor (LRF-1) because it is highly expressed in regenerating livers (23). Expression of ATF3 is known to modulate glucose homeostasis and other primary functions of the liver and likely plays a role in altering cell function before cell-cycle entry (24). Ets-2 is another immediate-early transcription factor that was highly induced after PHx. Ets-2, in conjunction with C/EBPα, and C/EBPβ, rapidly increases transcription from the p21 promoter via multiple binding elements within the enhancer region (25). Expression of Ets-2 has direct downstream effects on both cell-cycle progression and MAPK signaling.
We also observed that the gene encoding the peroxisome proliferator-activated receptor α (PPARα) was down-regulated during liver regeneration. PPAR genes mediate the proliferation of peroxisomes, which are organelles that participate in many primary functions of the liver including lipid metabolism, catabolism of purines, polyamines, and amino acids, H2O2-based respiration, cholesterol synthesis, and the production of bile acids (26). Down-regulation of PPAR linked to an immediate-early response to functional deficiency may be a natural response to the increase in H2O2 concentration in the remaining hepatocytes after PHx. Retinoic acid, a natural ligand of the retinoid X receptor (RXR), has been implicated as an important cytokine in regenerating livers (27). Consistent with a functional role of retinoic acid in liver regeneration, we observe the induction of Stra14 (stimulated by retinoic acid 14), the role of which in liver regeneration is unknown.
We also detected transient expression of other transcription factors such as hepatocyte nuclear factor 3(HNF-3)β and -γ, which are known to activate liver-specific genes such as albumin, and influence expression of genes involved in bile acid and glucose homeostasis (28, 29). HNF-3 isoforms mediate the hepatocyte-specific transcription of numerous genes important for liver function, and homozygous knockout mice do not survive embryonic development (28, 29). Gene expression of HNF-3 isoforms are reduced in the liver after injury by CCl4 (30), a model system for studying liver regeneration, suggesting an important role for these transcription factors in response to damage and to controlling the differentiation state and proliferation of hepatocytes.
By 240 min, we observed transcription factors associated with the beginnings of tissue remodeling. One example is the hypoxia-induced factor (HIF1α), which is an angiogenic gene expressed during oxygen starvation (hypoxia) that promotes vascular growth (31). At this time point, three IFN-inducible transcription factors also were induced: IFN-stimulated gene factor-3 (ISGF-3) and IFN regulatory factors-1 and -2 (IRF-1 and IRF-2). ISGF-3 and IRF-1 have been identified as transcriptional activators of IFN-β signaling, whereas IRF-2 is thought to act as a repressor of such activity (32). Given that IFN-β was induced by 90 min after PHx and this cytokine has been involved with cell-growth suppression (33, 34), these results suggest that ISGF-3, IRF-1, and IRF-2 may differentially modulate regeneration. Although many of the immediate-early transcription factors induced after PHx have proliferative functions, several genes involved in regulation and arrest of the cell cycle were also induced. This apparent dichotomy must be linked with the precise control of cell-cycle entry and progression.
Cell-Cycle Genes.
During hepatocyte priming in mice, we observed genes related to the cell cycle up-regulated as early as 10 min after PHx (Table 1). Consistent with the expression of both pro- and antiproliferative transcription factors, we observe the differential regulation of genes that stimulate and inhibit cell-cycle entry. Overall, we detected the differential regulation of 19 cell-cycle control genes during the time course of hepatocyte priming, and many are detected far earlier than reported previously. The majority of the genes are checkpoint genes at major cell-cycle transitions that can act to inhibit the cell cycle.
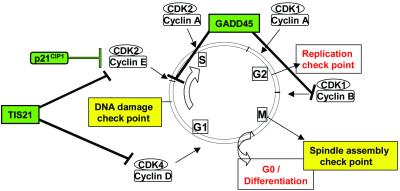
Three cell-cycle checkpoint genes induced during liver regeneration were GADD45, TIS21, and p21, which act at different cell-cycle transitions (Fig. 2). The levels of TIS21 mRNA, a p53-dependent growth arrest gene that inhibits the G1/S transition (35), increased by 10 min and reached maximum levels by 90–240 min after PHx (Table 1) as also confirmed by Northern blot (Fig. 1). Immediate-early transcription factors regulate the expression of the cell-cycle checkpoints (25), thus implying that autonomous control of cell-cycle entry by the hepatocytes begins almost immediately. In addition, we observe the up-regulation of cell-cycle genes that are apoptosis inhibitors such as Bcl-2, Bcl-X, and GADD45, previously implicated in hepatocyte priming (3).
Fig 2.
The influence of observed genes on cell cycle.
Signal Transduction.
We observed differential regulation of several genes related to signal transduction. For example, we observe the differential regulation of genes related to the activation of MAPK and related pathways. MAPK signaling is mediated by increased expression of the cyclin-dependent kinase inhibitor protein p21, which is induced by Ets-2 and C/EBP (25), all of which are observed in our gene-expression profile. MAPK activation has been linked to interleukin-6 (IL-6) pathways (8) and is an essential component of the genetic program that leads quiescent hepatocytes into a proliferative state. Our results with regards to MAPK-related pathways are consistent with the findings of Taub and coworkers, which are discussed in detail elsewhere (8).
It is noteworthy also that follistatin and inhibin, inhibitors of activin A, have proliferative properties and were up-regulated during hepatocyte priming. Activin A is a member of the transforming growth factor-β superfamily and is known to be involved in kidney organogenesis and development (36). Activin A can block the activity of the key inflammatory cytokine IL-6, which is known to be critical in liver regeneration (37). Up-regulation of inhibitors of activin A therefore are likely to play a role in hepatocyte priming by enhancing the effects of IL-6, which leads to proliferation.
Extracellular Matrix/Cell Structure- and Membrane-Associated Genes.
It is well established that modification of the extracellular matrix is integrally linked with liver regeneration (3). We have identified 28 differentially regulated genes that are associated with cell structure and extracellular matrix modification. Cyr61, an angiogenic extracellular matrix modifier (38, 39), appeared as early as 10 min after PHx and was overexpressed consistently during hepatocyte priming. Fisp12 (CTGF), another angiogenic extracellular matrix modifier that usually is observed in conjunction with Cyr61 (40), appeared only at the 90-min time point onward. The role of these two genes in liver regeneration has not been established previously, although they seem to be critical to the formation of new blood vessels and critical to proliferation in certain cancers (41). Both Cyr61 and Fisp12 are involved in wound healing and can stimulate chemotaxis and promote proliferation in endothelial cells and fibroblasts in culture and induce neovascularization in vivo (42). They also promote adhesive signaling responses that lead to sustained activation of p42/p44 MAPKs and prolonged gene-expression changes including up-regulation of MMP-1 (collagenase-l) and MMP-3 (stromelysin-l) mRNAs (42). The remainder of the genes began to be differentially expressed 90 min after PHx, suggesting that there is a lag between the initial response to PHx and extracellular modification. This was confirmed also by the fact that little or no induction of matrix metalloproteinases (MMP, proteinases known to play a role in tissue remodeling; ref. 43) was detected by microarray (Table 1) up to the 240-min time point.
It also is worth mentioning that plasminogen activator inhibitor 1 (PAI-1), a serine protease inhibitor that specifically inhibits plasmin activation (44) and liver regeneration, was very highly expressed from 90 min after PHx onward. PAI-1 specifically inhibits liver regeneration (3, 4) by forming a complex with the urokinase-type plasminogen activator (uPA) and contributes to the inactivation of HGF. HGF is a known stimulus of liver regeneration after priming (3, 4).
We also observed induction of RhoB, which regulates signal transduction from plasma membrane receptors (45, 46). RhoB is known also to regulate DNA synthesis and is expressed as a result of genotoxic stress (47). A number of other genes associated with cell adhesion and migration appear later in the priming phase (Table 1). Related to genes that are involved with cell adhesion and migration are those that are involved with cell–cell communication. Connexins comprise a class of cell membrane proteins that allow passive transport of small molecules between networked cells in tissues (48). Connexin 26, which shows antiproliferative behavior when overexpressed in human hepatoma cells (49), appeared up-regulated during liver regeneration. We also observed clcn3 down-regulated during the priming phase of hepatocyte proliferation. This gene is a voltage-gated chloride channel for regulation of cell volume (50). Expression of both pro- and antiproliferative extracellular matrix-modifying genes, exemplified by Cyr61 and Fisp12 compared with plasminogen activator inhibitor 1 and RhoB, is consistent with observations of expression patterns of transcription factors and cell-cycle genes and again suggestive of tight control of cell-cycle entry during hepatocyte priming.
Inflammatory Responses.
The pioneering work by Taub and coworkers regarding the role of IL-6 in liver regeneration in mice establishes the importance of inflammatory cytokines during hepatocyte priming (8). Inflammatory responses have been implicated in the priming of liver and other types of regeneration (51). For example, prevention of macrophage invasion impairs peripheral axonal regeneration, whereas implantation of macrophages into central nervous system nerves allows them to regenerate after axonal crush (51). Inflammation also is implicated in secondary degeneration after spinal cord injury (51). Inflammatory responses can be triggered by cytokines such as those of the TNF family, which activate immediate-early genes such as AP-1 (52). With the exception of IFN-β that, as mentioned before, was detected 90 min after PHx, other cytokines such as IL-6, IL-2, IL-3, IL-4, IL-5, TNF-α, TNF-β, and IFN-γ were not found in our samples, indicating that if induction of these genes occurred, it was below our detection limit. However, the messages for the chemoattractants MCP-1 and GRO were increased in the regenerating liver by 30 and 90 min after PHx, respectively. These chemokines can recruit monocytes/macrophages, which have the potential to exert both stimulatory and inhibitory influences on hepatocyte proliferation (53), in the liver after PHx (54). MCP-1 and GRO also may play a role in angiogenesis and tissue remodeling (55).
Other inflammatory genes included CD14, the lipopolysaccharide receptor, being highly induced after PHx, consistent with previous observations by Taub and coworkers (8), and the gibbon ape leukemia virus cell-surface receptor (glvr-1) that is involved in sodium/phosphate cotransport and induced during inflammatory responses (56). Up-regulation of the latter gene indicates an increase in the intracellular import of inorganic phosphate, which is required for activation of signaling pathways involving phosphorylation and nucleotide triphosphate synthesis. Northern blot analysis confirmed the increase in mRNA for the glvr-1 gene in the regenerating liver (Fig. 1).
Glucose- and Metabolism-Related Genes.
The liver plays an important role in maintaining metabolic and biosynthetic homeostasis. Glucose homeostasis is an important liver function and involves glycogenolysis, which breaks down glycogen into glucose, and gluconeogenesis, which involves synthesis of glucose from noncarbohydrate precursors (57). In a regenerating liver the majority of homeostatic responses involving metabolic functions occur after the priming phase of hepatocytes (15). In addition to previously mentioned genes ATF3 and PPARα, we observed the up-regulation of phosphoenol-pyruvate carboxykinase (PEPCK) and glucose-6-phosphatase (G6Pase), which are known to be involved in maintaining glucose levels after the acute loss of liver mass posthepatectomy (15, 57). G6Pase is involved in glycogenolysis and associated with the endoplasmic reticulum (ER), where it hydrolyses the G6P into glucose and phosphate. Misregulation of this gene has been implicated in type 1 glycogen storage diseases (58). PEPCK is a gene involved in the synthesis of glucose from noncarbohydrate precursors. In addition, we observed up-regulation of the insulin-like growth factor-binding protein (IGFBP-1), which is induced by IL-6 and HGF, and is known to be up-regulated during the course of liver regeneration (59). IGFBP-1 shares common promoter elements with other hepatic genes associated with the maintenance of metabolic homeostasis following large functional deficiency after PHx such as G6Pase (20).
Heme oxygenase 1 (HMOX1) is a protein that catalyzes the oxygen-dependent degradation of heme to biliverdin, free iron, and carbon monoxide. Of the two isoforms, the inducible HMOX1 primarily functions in the liver and the spleen. The increased expression of HMOX1 during liver regeneration is likely a reflection of the increased metabolic workload of the remaining hepatocytes after PHx and also may mediate oxidative stress by modulating iron levels and indirectly participate in antiproliferative proapoptotic pathways that control cell-cycle entry (60).
Uncharacterized Genes.
We have identified a number of genes corresponding to expressed sequence tags that were differentially expressed after PHx in mice. The molecular function can be inferred for these genes based on sequence homology. For example, the genes AA107455 and appear to have high homology to elongation factor-binding proteins. The gene appears homologous to a serine protease inhibitor, and W08822 is homologous to a putative GTPase-activating protein for Arf. However, sequence homology does not reveal the cellular or physiological role of these proteins, and their specific roles in liver regeneration are unclear.
Conclusions.
We have shown that the genome-wide expression profile of hepatocyte priming after PHx in mice is complex and covers different classes of proteins including transcription factors, metabolic enzymes, proteins associated with stress and inflammatory responses, and those involved in cytoskeletal and extracellular matrix modification. We have adapted a diagram originally produced by Fausto (3) to summarize these results (Fig. 3, which is published as supporting information on the PNAS web site, www.pnas.org). These genes are likely to have a broad effect on the liver. Along with the concept of hepatocyte priming (3, 10–12), it is worth mentioning that all these changes occurred well before DNA synthesis, suggesting that the transcriptional control of liver regeneration involves early and diverse cellular responses. It also was somewhat surprising to find that the expression levels of genes associated with the cell cycle over the time course of 4 h indicate that antiproliferative genes are favored during hepatocyte priming. These potential antiproliferative responses are in keeping with the concept of an autonomous control of cell-cycle entry by the hepatocytes and suggest that tight regulation of liver cell proliferation originates very early after a regenerative stimulus. Future experiments to determine which of the changes during hepatocyte priming are primary-cause factors in tissue regeneration may provide new insights into regenerative processes in mammals and potentially may lead to new approaches to the development of therapeutic agents for the treatment of liver diseases.
Supplementary Material
Acknowledgments
We thank Heike Mendez, Rick Koch, and Margie Chadwell for excellent technical assistance. We also thank Ian Campbell, Valerie Asensio, and Monte Hobbs for providing the cytokine, chemokine, and metalloproteinase gene probes used in the RPA experiments. This work was supported by National Institutes of Health Grants AI40696 (to L.G.G.) and CA40489 (to F.V.C.) and funds from Novartis. This is manuscript number 15024-CH from The Scripps Research Institute.
Abbreviations
PHx, partial hepatectomy
HGF, hepatocyte growth factor
TNF, tumor necrosis factor
RPA, RNase protection assay
MAPK, mitogen-activated protein kinase
C/EBP, CCAAT/enhancer-binding protein
PPAR, proliferator-activated receptor
References
- 1.Brockes J. P. (1997) Science 276, 81-87. [DOI] [PubMed] [Google Scholar]
- 2.Carlson B. M. (1998) Wound Repair Regen. 6, 425-433. [DOI] [PubMed] [Google Scholar]
- 3.Fausto N. (2000) J. Hepatol. 32, 19-31. [DOI] [PubMed] [Google Scholar]
- 4.Michalopoulos G. K. & DeFrances, M. C. (1997) Science 276, 60-66. [DOI] [PubMed] [Google Scholar]
- 5.Thorgeirsson S. S. (1996) FASEB J. 10, 1249-1256. [PubMed] [Google Scholar]
- 6.Sell S. (1994) Mod. Pathol. 7, 105. [PubMed] [Google Scholar]
- 7.Overturf K., al-Dhalimy, M., Ou, C. N., Finegold, M. & Grompe, M. (1997) Am. J. Pathol. 151, 1273-1280. [PMC free article] [PubMed] [Google Scholar]
- 8.Li W., Liang, X., Leu, J. I., Kovalovich, K., Ciliberto, G. & Taub, R. (2001) Hepatology 33, 1377-1386. [DOI] [PubMed] [Google Scholar]
- 9.Taub R., Greenbaum, L. E. & Peng, Y. (1999) Semin. Liver Dis. 19, 117-127. [DOI] [PubMed] [Google Scholar]
- 10.Webber E. M., Godowski, P. J. & Fausto, N. (1994) Hepatology 19, 489-497. [PubMed] [Google Scholar]
- 11.Fausto N., Laird, A. D. & Webber, E. M. (1995) FASEB J. 9, 1527-1536. [DOI] [PubMed] [Google Scholar]
- 12.Webber E. M., Bruix, J., Pierce, R. H. & Fausto, N. (1998) Hepatology 28, 1226-1234. [DOI] [PubMed] [Google Scholar]
- 13.Weglarz T. C. & Sandgren, E. P. (2000) Proc. Natl. Acad. Sci. USA 97, 12595-12600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cressman D. E., Diamond, R. H. & Taub, R. (1995) Hepatology 21, 1443-1449. [PubMed] [Google Scholar]
- 15.Taub R. (1996) FASEB J. 10, 413-427. [PubMed] [Google Scholar]
- 16.Guidotti L. G., Matzke, B., Schaller, H. & Chisari, F. V. (1995) J. Virol. 69, 6158-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pagenstecher A., Wussler, E. M., Opdenakker, G., Volk, B. & Campbell, I. L. (2001) J. Neuropathol. Exp. Neurol. 60, 598-612. [DOI] [PubMed] [Google Scholar]
- 18.Kakimi K., Lane, T. E., Wieland, S., Asensio, V. C., Campbell, I. L., Chisari, F. V. & Guidotti, L. G. (2001) J. Exp. Med. 194, 1755-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cressman D. E., Greenbaum, L. E., DeAngelis, R. A., Ciliberto, G., Furth, E. E., Poli, V. & Taub, R. (1996) Science 274, 1379-1383. [DOI] [PubMed] [Google Scholar]
- 20.Leu J. I., Crissey, M. A., Leu, J. P., Ciliberto, G. & Taub, R. (2001) Mol. Cell Biol. 21, 414-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inuzuka H., Nanbu-Wakao, R., Masuho, Y., Muramatsu, M., Tojo, H. & Wakao, H. (1999) Biochem. Biophys. Res. Commun. 265, 664-668. [DOI] [PubMed] [Google Scholar]
- 22.Chung K. C., Gomes, I., Wang, D. H., Lau, L. F. & Rosner, M. R. (1998) Mol. Cell. Biol. 18, 2272-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu J. C., Bravo, R. & Taub, R. (1992) Mol. Cell. Biol. 12, 4654-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allen-Jennings A. E., Hartman, M. G., Kociba, G. J. & Hai, T. (2001) J. Biol. Chem. 276, 29507-29514. [DOI] [PubMed] [Google Scholar]
- 25.Park J. S., Qiao, L., Gilfor, D., Yang, M. Y., Hylemon, P. B., Benz, C., Darlington, G., Firestone, G., Fisher, P. B. & Dent, P. (2000) Mol. Biol. Cell. 11, 2915-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kersten S., Desvergne, B. & Wahli, W. (2000) Nature (London) 405, 421-424. [DOI] [PubMed] [Google Scholar]
- 27.Imai T., Jiang, M., Kastner, P., Chambon, P. & Metzger, D. (2001) Proc. Natl. Acad. Sci. USA 98, 4581-4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rausa F. M., Tan, Y. J., Zhou, H. P., Yoo, K. W., Stolz, D. B., Watkins, S. C., Franks, R. R., Unterman, T. G. & Costa, R. H. (2000) Mol. Cell. Biol. 20, 8264-8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaestner K. H. (2000) Trends Endocrinol. Metab. 11, 281-285. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura T., Akiyoshi, H., Shiota, G., Isono, M., Nakamura, K., Moriyama, M. & Sato, K. (1999) FEBS Lett. 459, 1-4. [DOI] [PubMed] [Google Scholar]
- 31.Zhu H. & Bunn, H. F. (2001) Science 292, 449-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harada H., Taniguchi, T. & Tanaka, N. (1998) Biochimie 80, 641-650. [DOI] [PubMed] [Google Scholar]
- 33.Romeo G., Fiorucci, G., Chiantore, M. V., Percario, Z. A., Vannucchi, S. & Affabris, E. (2002) J. Interferon Cytokine Res. 22, 39-47. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka N. & Taniguchi, T. (2000) Semin. Cancer Biol. 10, 73-81. [DOI] [PubMed] [Google Scholar]
- 35.Guardavaccaro D., Corrente, G., Covone, F., Micheli, L., D'Agnano, I., Starace, G., Caruso, M. & Tirone, F. (2000) Mol. Cell. Biol. 20, 1797-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maeshima A., Nojima, Y. & Kojima, I. (2001) Cytokine Growth Factor Rev. 12, 289-298. [DOI] [PubMed] [Google Scholar]
- 37.Russell C. E., Hedger, M. P., Brauman, J. N., de Kretser, D. M. & Phillips, D. J. (1999) Mol. Cell. Endocrinol. 148, 129-136. [DOI] [PubMed] [Google Scholar]
- 38.Babic A. M., Kireeva, M. L., Kolesnikova, T. V. & Lau, L. F. (1998) Proc. Natl. Acad. Sci. USA 95, 6355-6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grzeszkiewicz T. M., Kirschling, D. J., Chen, N. & Lau, L. F. (2001) J. Biol. Chem. 276, 21943-21950. [DOI] [PubMed] [Google Scholar]
- 40.Babic A. M., Chen, C. C. & Lau, L. F. (1999) Mol. Cell. Biol 19, 2958-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie D., Miller, C. W., O'Kelly, J., Nakachi, K., Sakashita, A., Said, J. W., Gornbein, J. & Koeffler, H. P. (2001) J. Biol. Chem. 276, 14187-14194. [DOI] [PubMed] [Google Scholar]
- 42.Chen C. C., Mo, F. E. & Lau, L. F. (2001) J. Biol. Chem. 276, 47329-47337. [DOI] [PubMed] [Google Scholar]
- 43.Sternlicht M. D. & Werb, Z. (2001) Annu. Rev. Cell Dev. Biol. 17, 463-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimizu M., Hara, A., Okuno, M., Matsuno, H., Okada, K., Ueshima, S., Matsuo, O., Niwa, M., Akita, K., Yamada, Y., et al. (2001) Hepatology 33, 569-576. [DOI] [PubMed] [Google Scholar]
- 45.Kato M., Iwamoto, H., Higashi, N., Sugimoto, R., Uchimura, K., Tada, S., Sakai, H., Nakamuta, M. & Nawata, H. (1999) J. Hepatol. 31, 91-99. [DOI] [PubMed] [Google Scholar]
- 46.Fritz G., Gnad, R. & Kaina, B. (1999) Anticancer Res. 19, 1681-1688. [PubMed] [Google Scholar]
- 47.Kovalovich K., DeAngelis, R. A., Li, W., Furth, E. E., Ciliberto, G. & Taub, R. (2000) Hepatology 31, 149-159. [DOI] [PubMed] [Google Scholar]
- 48.Hand G. M., Muller, D. J., Nicholson, B. J., Engel, A. & Sosinsky, G. E. (2002) J. Mol. Biol. 315, 587-600. [DOI] [PubMed] [Google Scholar]
- 49.Muramatsu A., Iwai, M., Morikawa, T., Tanaka, S., Mori, T., Harada, Y. & Okanoue, T. (2002) Carcinogenesis 23, 351-358. [DOI] [PubMed] [Google Scholar]
- 50.Lamb F. S., Clayton, G. H., Liu, B. X., Smith, R. L., Barna, T. J. & Schutte, B. C. (1999) J. Mol. Cell. Cardiol. 31, 657-666. [DOI] [PubMed] [Google Scholar]
- 51.Piehl F. & Lidman, O. (2001) Immunol. Rev. 184, 212-225. [DOI] [PubMed] [Google Scholar]
- 52.Kyriakis J. M. (1999) Gene Expression 7, 217-231. [PMC free article] [PubMed] [Google Scholar]
- 53.Takeishi T., Hirano, K., Kobayashi, T., Hasegawa, G., Hatakeyama, K. & Naito, M. (1999) Arch. Histol. Cytol. 62, 413-422. [DOI] [PubMed] [Google Scholar]
- 54.Bouwens L., Baekeland, M. & Wisse, E. (1984) Hepatology 4, 213-219. [DOI] [PubMed] [Google Scholar]
- 55.Rossi D. & Zlotnik, A. (2000) Annu. Rev. Immunol. 18, 217-243. [DOI] [PubMed] [Google Scholar]
- 56.Mansfield K., Teixeira, C. C., Adams, C. S. & Shapiro, I. M. (2001) Bone (NY) 28, 1-8. [DOI] [PubMed] [Google Scholar]
- 57.Haber B. A., Chin, S., Chuang, E., Buikhuisen, W., Naji, A. & Taub, R. (1995) J. Clin. Invest. 95, 832-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lei K. J., Chen, H. W., Pan, C. J., Ward, J. M., Mosinger, B., Lee, E. J., Westphal, H., Mansfield, B. C. & Chou, J. Y. (1996) Nat. Genet. 13, 203-209. [DOI] [PubMed] [Google Scholar]
- 59.Lee J., Greenbaum, L., Haber, B. A., Nagle, D., Lee, V., Miles, V., Mohn, K. L., Bucan, M. & Taub, R. (1994) Hepatology 19, 656-665. [DOI] [PubMed] [Google Scholar]
- 60.Thom S. R., Fisher, D., Xu, Y. A., Notarfrancesco, K. & Ischiropoulos, H. (2000) Proc. Natl. Acad. Sci. USA 97, 1305-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.