Abstract
Caveolin-1 was the first protein identified that colocalizes with the ≈10-nm filaments found on the inside surface of caveolae membranes. We have used a combination of electron microscopy (EM), circular dichroism, and analytical ultracentrifugation to determine the structure of the oligomers that form when the first 101 aa of caveolin-1 (Cav1–101) are allowed to associate. We determined that amino acids 79–96 in this caveolin-1 fragment are arranged in an α-helix. Cav1–101 oligomers are ≈11 nm in diameter and contain seven molecules of Cav1–101. These subunits, in turn, are able to assemble into 50 nm long × 11 nm diameter filaments that closely match the morphology of the filaments in the caveolae filamentous coat. We propose that the heptameric subunit forms in part through lateral interactions between the α-helices of the seven Cav1–101 units. Caveolin-1, therefore, appears to be the structural molecule of the caveolae filamentous coat.
A unique membrane coat is found on the inside surface of fibroblast (1) and endothelial cell (2) caveolae. Each coat consists of 4–6, concentrically arranged, 10-nm-wide filaments that are embedded in the membrane. Sometimes the filaments are curved or spiral, whereas other times they run parallel to each other. This filamentous coat decorates both planar and deeply invaginated regions of membrane. The full extent of the coat is best appreciated, however, in the planar coats, where they can measure up to 125 nm in diameter. This coat is not removed by washing the membrane with high-salt solutions, suggesting it is not constructed from peripheral membrane proteins. By contrast, treatment of the membrane with cholesterol-binding drugs like nystatin or filipin cause the coat to disassemble. During disassembly, the filaments disintegrate into particles that measure ≈10 nm in diameter (1).
The only known protein component of the caveolae coat is caveolin-1, which has been localized by immunogold to individual filaments in the coat (1). There are three caveolin genes (designated 1, 2, and 3), coding for five distinct proteins (3). Each share in common a long hydrophobic region, which is probably inserted in the membrane, and N-terminal and C-terminal portions that most likely are in the cytoplasm. Whether caveolin-1 is the structural subunit of the filament has not been determined.
The apparent molecular mass of caveolin-1 is 22–24 kDa. Therefore, a 10-nm-wide filament made from pure caveolin-1 would most likely be constructed from caveolin-1 oligomers. The ability of caveolin-1 to oligomerize is well documented. There appear to be two aspects to the oligomerization process. A glutathione S-transferase (GST)-fusion peptide of the N-terminal cytoplasmic portion of caveolin-1 (amino acids 1–101) spontaneously forms oligomers in vitro (4). Removing amino acids 81–101 from the peptide abolishes oligomerization. Oligomerization of the full-length protein, on the other hand, appears to be stabilized by the palmitoylation of cysteine residues located at positions 133, 143, and 156 (5). Indeed, deletion of amino acids 134–154 impairs oligomerization as well as transport of caveolin-1 from the Golgi apparatus to the cell surface (6). Exactly how oligomeric caveolin-1 forms the filamentous caveolae coat, however, cannot be determined without high-resolution structural information.
Materials and Methods
Construction of Caveolin Bacterial Expression Systems.
All caveolin constructs were generated by PCR from cDNAs encoding the indicated caveolin isoform by using the Expand High Fidelity PCR system (Roche Applied Science, Indianapolis). The PCR products were subcloned into the multicloning site (BamHI–EcoRI) of the vector pGEX-KT. Plasmids were constructed to encode different caveolin-1, caveolin-2, and caveolin-3 N-terminal cytoplasmic fragments. To prepare a caveolin-1 mutant corresponding to the caveolin-3 mutation found in patients with limb-girdle muscular dystrophy, 1C, the plasmid encoding caveolin-1 wild-type 1–101 was mutagenized by using the QuickChange site-directed mutagenesis protocol (Stratagene) to delete amino acids 91–93 (TFT). The 66A70 mutant Cav1–101 was described (6). EcoRI and BamHI restriction sites were added to the PCR products of all constructs and subcloned into pGEX-KT vectors. Caveolin-1 fragment 80–101 was synthesized by solid-phase methods, purified by HPLC, and characterized by mass spectrometry (MS). Escherichia coli (BL21 strain, Novagen) were transfected with pGEX-KT plasmids, and expression was induced by the addition of 0.1 mM isopropyl β-d-thiogalactopyranoside (IPTG; Sigma) at 25°C overnight. Cells were harvested, resuspended in PBS, and lysed by passing three times through an EmulsiFlex-C5 cell disrupter (Avestin, Ottawa) at 14,000 psi (1 psi = 6.9 kPa). Supernatants were separated from insoluble pellet by spinning the lysate at 28,000 × g for 30 min. The supernatants were then incubated with a slurry of glutathione agarose beads (Sigma). The beads were extensively washed with PBS until there was no detectable UV absorption in the eluate. The sample was equilibrated with buffer A (50 mM Tris, pH 8.0/0.2 M NaCl/2.5 mM CaCl2) before thrombin (12 units/liter; Sigma) was added and the sample incubated for 1.5 h at 25°C. Cleaved products were eluted and further purified by anion-exchange chromatography on MonoQ (Amersham Biosystems, Piscataway, NJ).
Column Chromatography.
Purified samples of the indicated recombinant protein (0.2–5.0 mg/ml) were analyzed by using a HiLoad 16/60 Superdex 75 or on a Superdex 200 gel filtration column (Amersham Pharmacia Biotech). The column was preequilibrated with buffer B (200 mM sodium phosphate/300 mM NaCl, pH 6.3) and calibrated with standard proteins of known molecular size (Amersham Pharmacia Biotech). Samples were eluted at a flow rate of 1 ml/min and the fractions were monitored by absorbance at 280 nm.
Analytical Ultracentrifugation.
Analytical ultracentrifugation experiments were performed by using a Beckman XL-I analytical ultracentrifuge. For sedimentation equilibrium experiments, samples were loaded in an An60Ti rotor with an equilibrium six-channel centerpiece (path length 1.2 cm) and run at 9,000, 12,000, and 15,000 rpm, at 4°C. Data were collected at a wavelength of either 280 or 305 nm. Background absorbance was estimated by overspeeding at 42,000 rpm until a flat baseline was obtained. Analysis of the data, including estimation of average molecular weight as a function of protein concentration (Fig. 4A), was carried out by using Beckman software. The calculated molecular weight of Cav11–101 monomers was 11,725. Partial specific volumes, estimated by using the SEDIMENTATION INTERPRETATION PROGRAM V.1.03 (Biomolecular Interaction Technologies Center, Durham, NH) based on the method of Cohn and Edsall (7), were 0.7209 at 4°C and 0.7277 at 20°C. Extinction coefficients were calculated as 19,060 M−1⋅ cm−1 at 280 nm by the method of Gill and von Hippel (8). Sedimentation velocity data were collected at 280 nm, 20°C, and 40,000 rpm by using the 12-mm charcoal-filled double-sector centerpiece. Data were analyzed by using the g(s*) method in the Beckman software. In this method, an apparent sedimentation coefficient distribution function [g(s*)] is calculated from the absorbance curves collected at successive intervals (9, 10). Molecular weights (M) and diffusion coefficients (D) were calculated by using the relations s = M(1−νρ)/Naf and D = kT/f, where s is the sedimentation coefficient, f is the frictional coefficient, T is the absolute temperature, k is Boltzmann's constant, ν is the partial specific volume, ρ is the solvent density, and NA is Avogadro's number. Hydrodynamic radii (R) were calculated by using the Stokes equation: f = 6πηR.
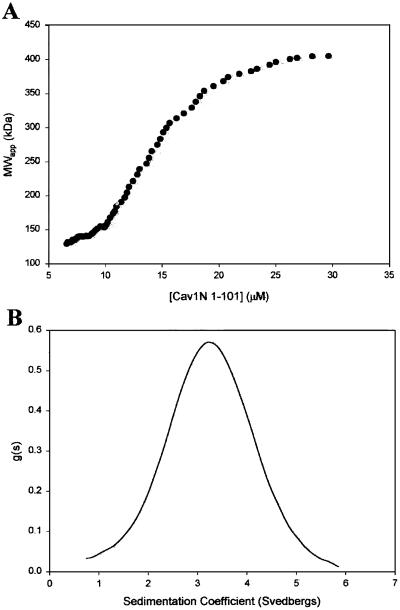
Fig 4.
Analytical ultracentrifugation suggests that Cav11–101 self-assembles into at least two oligomeric states. (A) Sedimentation equilibrium analysis. Cav11–101 peptides were purified as described. Various concentrations of the peptide were subjected to equilibrium centrifugation at various speeds in an An60Ti rotor. The plot shows the distribution of average molecular weights as a function of peptide concentration calculated from multiple runs at 9,000 rpm. At this speed, molecular weights of 400,000–450,000 were consistently observed at the highest concentrations, near the bottom of the centrifuged cell. However, the average molecular weight of Cav11–101 oligomers at each concentration decreased progressively with increasing rotor speed (not shown), suggesting pressure-dependent dissociation. (B) Sedimentation velocity analysis. The symmetric nature of the sedimentation coefficient distribution function is indicative of a relatively homogenous population of oligomers. Thus, at the high speed used (55,000 rpm in the experiment shown), Cav11–101 has presumably undergone pressure-induced dissociation to a stable oligomeric state. Calculations based on sedimentation and diffusion constants obtained here suggest that this stable oligomer has a molecular weight of approximately 83,000.
Negative Staining.
Samples of the indicated caveolin peptide (5 μl at 0.5 mg/ml) were processed for negative staining by using glow-discharged, Formvar-coated, 400-mesh nickel grids. We used 1% aqueous uranyl acetate to stain all samples before viewing with the JEOL 1200 CX electron microscope at 80 kV.
Circular Dichroism.
The indicated caveolin peptide (20 μM) purified by FPLC was analyzed on an Aviv spectrophotometer by using a 0.01-cm pathlength cuvette at 25°C.
Results and Discussion
We have used biophysical techniques to obtain secondary and tertiary structural information about recombinant N-terminal fragments of caveolin-1 in solution. Wild-type and mutant versions of Cav11–101, Cav21–73, and Cav31–74 fused to glutathione S-transferase (GST) were expressed in E. coli BL21, purified by using a glutathione column, recovered after thrombin cleavage, and further purified by anion-exchange chromatography.
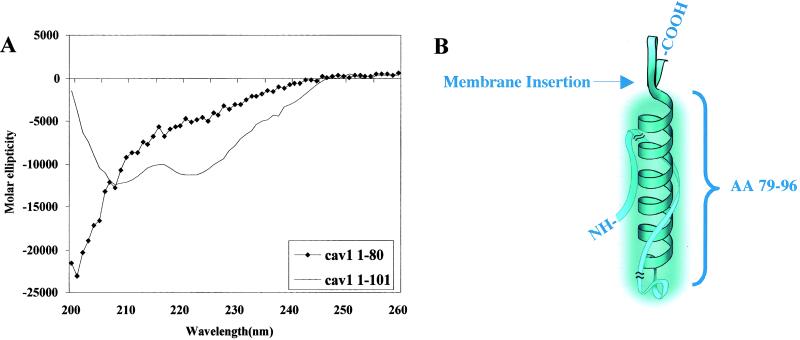
The secondary structure of wild-type Cav11–101 was determined by using circular dichroism (Fig. 1A). The characteristic double minima at 208 and 222 nm indicate that the peptide chain has an α-helical region. We estimate from the intensity of the minima that ≈20% of the peptide is helical. Tests of Cav21–73 and Cav31–74 showed that these molecules contained ≈25% α-helix (data not shown). The predictprotein program (EMBL-Heidelberg) suggests that the amino acid sequence from 79 to 96 in Cav11–101 is the most likely sequence to be in an α-helix. In agreement with this prediction, Cav11–80 did not display any 208/222 nm minima (♦). On the other hand, a peptide lacking the first 59 aa of Cav11–101 was ≈90% α-helical (data not shown). We used a helical wheel to visualize the region encompassing residues 79–96. An interesting feature to emerge from this representation is that on one side of the helix is an interdigitating set of basic and aromatic residues whereas the other side is made up of polar, uncharged residues. The charged residues spiral up the helix. Therefore, residues 79–96 form an α-helix and appear to be the only part of the N terminus with secondary structure. Fig. 1B shows a model that depicts how the helix is positioned in relation to the putative membrane insertion site. The model shows the sequence between 1 and 79 interacting with the helix (see below), but the exact organization of this region of the molecule is not known.
Fig 1.
Residues 79–96 in Cav11–101 form an α-helix. The indicated caveolin-1 peptides were prepared as described in the text. (A) Circular dichroism spectra of Cav11–101 (solid line) and Cav11–80 (♦). (B) A model of the caveolin-1 α-helix between amino acids 79–96. Shown wrapping around the helix is the region of the molecule between amino acids 1–80. Portions at either end of this region are truncated (indicated by slash marks) because of their uncertain location. We believe this part of the molecule contributes to the size of the unit, as indicated by the cloud surrounding the helix.
Caveolin Peptides Form Filaments.
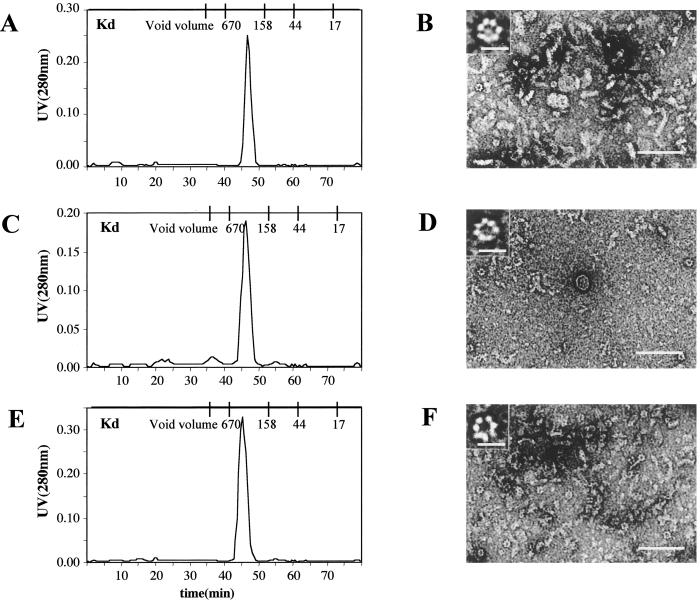
Gel filtration previously showed that Cav11–101 migrates anomalously on the Superdex 75 column (4). Rather than behaving like an 8 to12-kDa peptide, Cav11–101 elutes with an apparent molecular weight of ≈450,000 (Fig. 2A). We did not detect any protein in the region of the column that would correspond to monomeric Cav11–101. Gel electrophoresis of the peak fraction, however, showed that it was pure Cav11–101 (data not shown). A similar result was obtained when we performed gel filtration of Cav21–73 (Fig. 2C) and Cav31–74 (Fig. 2E). These results suggest that the N-terminal cytoplasmic portion of all three caveolin isoforms spontaneously form oligomers in solution with an apparent uniform size distribution.
Fig 2.
Gel filtration (A, C, and E) and negative staining (B, D, and F) analysis of Cav11–101, Cav21–73, and Cav31–74 structure. The indicated peptides were purified and applied to a Superdex 75 column. Shown are the elution profiles for Cav11–101 (A), Cav21–73 (C), and Cav31–74 (E). Peak fractions from each column were prepared for negative-staining electron microscopy (EM) as described. Representative images for Cav11–101 (B), Cav21–73 (D), and Cav31–74 (F) are shown. (Scale bars: B, D, and F, 0.1 μm; Inset, 0.01 μm.)
We used negative-staining electron microscopy (EM) to visualize the peak fractions for each caveolin isoform. In the case of Cav11–101 (Fig. 2B), we found that this fraction contained numerous, short filaments measuring 11 nm wide × ≈50 nm long. Scattered among the filaments were ring-shaped structures that, at higher magnification (see Inset), appeared to be composed of ≈7 subunits. Each ring measured ≈10 nm in diameter and in some images appeared to originate from the filaments. Cav21–73 (D) and Cav31–74 (F) oligomers were also filamentous in negative stain images, although they tended to be thinner (10 nm × 50 nm) than the Cav11–101 oligomers. The insets show examples of ring structures that were in these fractions. These images suggest to us that the caveolin oligomers eluting from the Superdex column are filaments composed of approximately five 10-nm diameter subunits. Each subunit, in turn, is composed of ≈7 copies of the monomeric N-terminal cytoplasmic tail of the indicated caveolin.
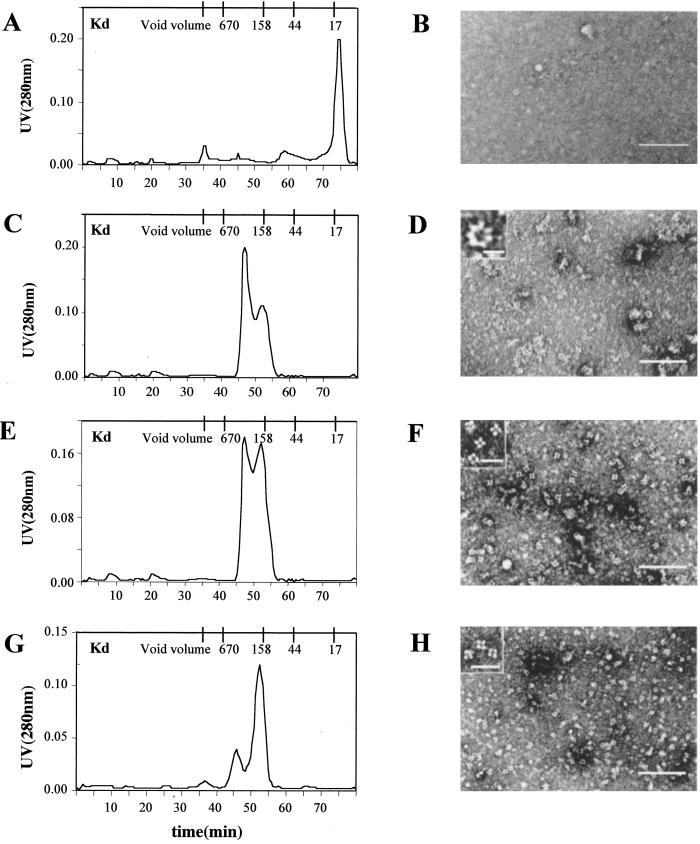
Most likely it is the α-helical region of the caveolin N-terminal cytoplasmic tail that mediates subunit assembly. Therefore, we prepared Cav11–80 and ran it on a gel filtration column (Fig. 3A). The peptide migrated on the column with an apparent molecular weight of <17,000, which suggests that it does not oligomerize. There was no evidence of filaments or rings in the negative stain images of these fractions (Fig. 3B). A very similar result was obtained by substituting alanine residues for amino acids 96 to 100 (data not shown).
Fig 3.
Gel filtration (A, C, E, and G) and negative staining (B, D, F, and H) analysis of Cav11–80, Cav11–101(Δ91–93), Cav160–101, and Cav11–101(66A70). The indicated peptides were purified and applied to a Superdex 75 column. Shown are the elution profiles of Cav11–80 (A), Cav11–101(Δ91–93) (C), Cav160–101 (E), and Cav11–101(66A70) (G). Peak fractions from each column were prepared for negative-staining EM as described. Representative images for Cav11–80 (B), Cav11–101(Δ91–93) (D), Cav160–101 (F), and Cav11–101(66A70) (H) are shown. (Scale bars: B, D, F, and H, 0.1 μm; Inset, 0.01 μm.)
This result prompted us to examine how various mutations in the Cav11–101 peptide affected oligomer formation. A three amino acid deletion (TFT) in caveolin-3 (Δ64–66) has been reported to cause limb-girdle muscular dystrophy 1C (LGMD-1C) (11, 12). We first looked at the effects of introducing the LGMD-1C (Δ91–93) mutation into Cav11–101 (Fig. 3C). The mutated peptide formed oligomers that by gel filtration appeared to be markedly smaller and more heterogeneous in size. Very few filaments were evident in negative stained images of the peak fractions, but numerous, normal appearing rings were present (Fig. 3D). At high magnification (Inset Fig. 3D), the Cav11–101(Δ91–93) rings were indistinguishable from wild-type Cav11–101 rings. The Cav11–101(Δ91–93) peptide, therefore, behaves as if it can still form rings, but these subunits are impaired in filament formation so that any filaments that form tend to disassemble during the preparation of the negative stain. By contrast, either deletion of the first 60 aa of Cav11–101 (Fig. 3 E and F) or substituting alanine residues at positions 66–70 (Fig. 3 G and H) resulted in the formation of tetrameric subunits but no filaments. Most likely, the tetramers corresponded to improperly assembled rings.
Analysis of Cav11–101 Oligomers by Analytical Centrifugation.
The EM results strongly suggest that the N-terminal, cytoplasmic portion of all caveolin isoforms spontaneously oligomerize to form ring-shaped structures consisting of seven copies of Cav11–101. These subunits, in turn, assemble into filaments ≈50 nm long. We used analytical ultracentrifugation to verify these results.
Analytical centrifugation is a method that could potentially be used to estimate the mass (by sedimentation equilibrium) and shape (by sedimentation velocity) of Cav11–101 oligomers. On reaching equilibrium, single homogeneous species form a concentration gradient from the meniscus to the bottom of the centrifuge cell, which gives a single straight line when plotted as the natural log of the concentration vs. radius2. The slope of this line can be used to calculate molecular weight. When Cav11–101 was centrifuged to equilibrium at three different rotor speeds, it became evident that multiple oligomeric species were present. For example, data obtained at 9,000 rpm did not define a single exponential concentration curve, and a replot of the data showed an upward curvature, indicative of self-association. By estimating the average molecular weight at each radial position in the centrifuge cell (using small subsets of the data centered at each position), a plot of molecular weight vs. Cav11–101 concentration was generated (Fig. 4A). At the highest protein concentrations, the molecular weights level off between 400,000 and 450,000, indicating that the largest oligomeric species is close to this molecular weight. Interestingly, as rotor speeds were increased, the oligomers at each concentration became progressively smaller, a phenomenon consistent with pressure-induced dissociation (13).
We next used the sedimentation velocity method to determine the hydrodynamic properties of Cav11–101. Sedimentation velocity experiments are carried out at high rotor speeds (40,000 and 55,000 rpm in our case) and, hence, under high pressures. Therefore, we anticipated that the filaments would disassemble to an even greater extent than in the low-speed sedimentation equilibrium runs. At both speeds we obtained reasonably symmetric sedimentation coefficient distribution profiles, consistent with the existence of a single homogenous species with a sedimentation coefficient of 3.25 S (Fig. 4B). Based on this value, and on the character of the distribution profile, it was possible to estimate a diffusion constant for the sedimenting particle of 3.25 × 10−7 cm2⋅s−1. Taken together, the sedimentation and diffusion constants suggest that the pressure-dissociated oligomer has a molecular weight of approximately 83,000 and a diameter of 12.2 nm. Assuming that this oligomer represents the subunit that assembles into filaments, we conclude that the particle consists of seven 11.75-kDa Cav11–101 monomers. This measurement matches well the dimensions of the heptameric unit seen by negative-stain EM.
Heptameric Cav11–101 and Caveolae Filaments.
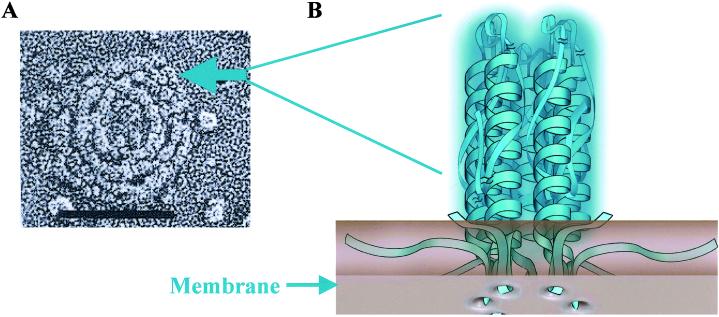
The particulate substructure of the caveolae coat filament was first noted in rapid-freeze deep-etch images of fibroblast membranes that had been treated with the cholesterol binding drug nystatin (arrow, Fig. 5A). These particles measure ≈10 nm in diameter, which is close to the diameter of the Cav11–101 ring (Inset, Fig. 2B). If the rings seen in negative staining correspond to the particles seen in rapid-freeze deep-etch imaging, then each particle is an oligomer consisting of seven Cav1 molecules assembled through lateral interactions between the Cav179–96 α-helices (Fig. 5B). Because Cav2 and Cav3 also form rings (Fig. 2 D and F), a particle potentially could contain any combination of caveolin isoforms. We conclude that a heptameric oligomer of caveolin is the basic building block of the caveolae coat filament.
Fig 5.
A model showing the relationship between the caveolae filament subunits seen in rapid-freeze deep-etch images (A) and the Cav1 heptameric structure seen by negative-stain EM (B). (A) Rapid-freeze deep-etch image of the caveolae filamentous coat in membranes that have been treated with nystatin to cause coat disassembly (1). The arrow points to one of numerous subunits that appear in regions where filament disassembly is occurring. These subunits measure ≈10 nm in diameter. (B) We believe that each of these subunits corresponds to a heptameric ring composed of seven caveolin-1 monomers joined through lateral interactions between their α-helices and associated parts of the molecule. A prediction made by this model is that the membrane insertion region of each α-helix will extend into the membrane to form of a ring of hydrophobic peptide loops. (Scale bar: 0.1 μm.)
What remains uncertain is the location of the first 78 aa in Cav1–101. The size of the individual subunits in the heptamer is larger than the size of an α-helix, which suggests that nonhelical regions of Cav101 may contribute to the structure of the subunit even though this region appears to have little secondary structure. Consistent with this conclusion, we found that removing the first 60 aa of Cav11–101 or changing amino acids 66–70 (IDFED) to alanine dramatically affected subunit assembly. We speculate that the acidic patch of amino acids between 67 and 70 may bind to the basic residues that spiral up the helix, thereby causing the tail to wrap around the helix (see model, Fig. 1B).
We also found that a three-amino-acid deletion (Δ91–93) in the upper region of the Cav1 helix is permissive for ring formation, but the rings are impaired in filament assembly. Besides validating the methods we used to monitor the assembly of caveolin filaments, these results suggest that ring and filament formation are complex and involve both hydrophobic and ionic interactions. The finding that salt concentration had little effect on filament stability (data not shown) supports this conclusion.
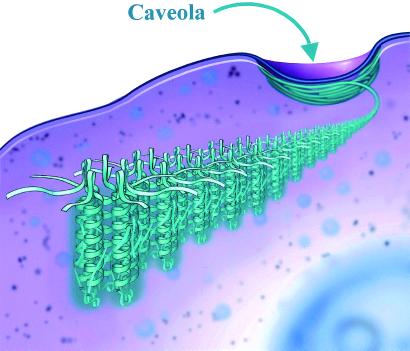
Most likely the membrane-insertion segment and C-terminal tail have little to do with heptamer assembly, but may profoundly influence filament formation (Fig. 6). The caveolin-1 α-helix (amino acids 79–96) is juxtaposed to the 33-aa long (starting at amino acid 101) hydrophobic loop that inserts the molecule into the membrane. Therefore, each helix in the heptamer has a hydrophobic loop extending into the membrane (Fig. 5B). In addition, two to three covalently attached palmitate residues at the C-terminal end of the membrane insertion region are part of the membrane anchor. The palmitate residues may be critical for the interaction of caveolin-1 with cholesterol (14). It is easy to appreciate from this model, therefore, how membrane lipids such as cholesterol might control filament assembly by increasing the stability of lateral interactions between the ring subunits.
Fig 6.
A model showing how caveolin-1 ring subunits assemble to make the 10-nm-thick filament of the caveolae coat.
Even at the relatively low resolution of these studies, there is nothing about the structure of caveolin to suggest that it functions directly in shaping caveolae membranes. Instead, we speculate that caveolin-1 plays a role in caveolae-lipid organization. The caveolin-1 ring subunit bears a striking resemblance to α-hemolysin oligomers (15). α-Hemolysin is a 33-kDa protein secreted by bacteria in a soluble form that oligomerizes into heptameric rings measuring 10 nm × 10 nm when it binds to erythrocytes. These rings form pores in the membrane, a process that depends on membrane cholesterol and sphingolipids. The primary sequences of caveolin-1 and α-hemolysin do not show any homology. This raises the possibility there might be other proteins in cells that function like caveolin-1 but cannot be recognized on the basis of their primary sequence.
Acknowledgments
We thank Heather Webster for her valuable technical assistance, Dr. Derk Binns for help with the analytical ultracentrifugation experiments, and Brenda Pallares for administrative assistance. We also thank Dr. Jose Rizo-Rey for his helpful suggestions. This work was supported by National Institutes of Health Grants HL 20948 and GM 52016, the Muscular Dystrophy Association, and the Perot Family Foundation.
Abbreviations
EM, electron microscopy
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Rothberg K. G., Heuser, J. E., Donzell, W. C., Ying, Y. S., Glenney, J. R. & Anderson, R. G. (1992) Cell 68, 673-682. [DOI] [PubMed] [Google Scholar]
- 2.Peters K. R., Carley, W. W. & Palade, G. E. (1985) J. Cell Biol. 101, 2233-2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smart E. J., Graf, G. A., McNiven, M. A., Sessa, W. C., Engelman, J. A., Scherer, P. E., Okamoto, T. & Lisanti, M. P. (1999) Mol. Cell. Biol. 19, 7289-7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sargiacomo M., Scherer, P. E., Tang, Z., Kubler, E., Song, K. S., Sanders, M. C. & Lisanti, M. P. (1995) Proc. Natl. Acad. Sci. USA 92, 9407-9411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monier S., Dietzen, D. J., Hastings, W. R., Lublin, D. M. & Kurzchalia, T. V. (1996) FEBS Lett. 388, 143-149. [DOI] [PubMed] [Google Scholar]
- 6.Machleidt T., Li, W. P., Liu, P. & Anderson, R. G. W. (2000) J. Cell Biol. 148, 17-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohn E. J. & Edsall, J. J., (1943) Proteins, Amino Acids, and Peptides as Ions and Dipolar Ions (Reinhold, New York).
- 8.Gill S. C. & von Hippel, P. H. (1989) Anal. Biochem. 182, 319-326. [DOI] [PubMed] [Google Scholar]
- 9.Stafford W. F. (1992) Anal. Biochem. 203, 295-301. [DOI] [PubMed] [Google Scholar]
- 10.Stafford W. F. (1994) Methods Enzymol. 240, 478-501. [DOI] [PubMed] [Google Scholar]
- 11.Galbiati F., Engelman, J. A., Volonte, D., Zhang, X. L., Minetti, C., Li, M., Hou, H., Jr., Kneitz, B., Edelmann, W. & Lisanti, M. P. (2001) J. Biol. Chem. 276, 21425-21433. [DOI] [PubMed] [Google Scholar]
- 12.Kunkel L. (1999) J. Child Neurol. 14, 33-34. [PubMed] [Google Scholar]
- 13.Harrington W. F. & Kegeles, G. (1973) Methods Enzymol. 27, 306-345. [DOI] [PubMed] [Google Scholar]
- 14.Uittenbogaard A. & Smart, E. J. (2000) J. Biol. Chem. 275, 25595-25599. [DOI] [PubMed] [Google Scholar]
- 15.Song L., Hobaugh, M. R., Shustak, C., Cheley, S., Bayley, H. & Gouaux, J. E. (1996) Science 274, 1859-1866. [DOI] [PubMed] [Google Scholar]