Abstract
Plants are attacked by a broad array of herbivores and pathogens. In response, plants deploy an arsenal of defensive traits. In Brassicaceae, the glucosinolate–myrosinase complex is a sophisticated two-component system to ward off opponents. However, this so-called “mustard oil bomb” is disarmed by a glucosinolate sulfatase of a crucifer specialist insect, diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae). Sulfatase activity of this enzyme largely prevents the formation of toxic hydrolysis products arising from this plant defense system. Importantly, the enzyme acts on all major classes of glucosinolates, thus enabling diamondback moths to use a broad range of cruciferous host plants.
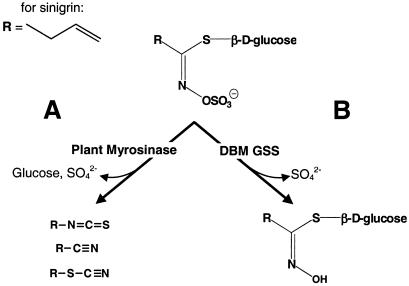
In response to biotic challenges, plants have evolved a broad variety of defense mechanisms. These include preformed physical and chemical barriers, as well as inducible defenses. A well-studied example is the glucosinolate–myrosinase system (Fig. 1A), also referred to as “the mustard oil bomb” (1, 2). Cruciferous plants synthesize glucosinolates, a class of plant secondary compounds that share a core consisting of a β-thioglucose moiety and a sulfonated oxime, but differ by a variable side chain derived from one of several amino acids (3). More than 100 different glucosinolates have been identified (4), and this diversity is believed to be of ecological importance (5–7). However, intact glucosinolates have limited biological activity. Their potency arises when plant tissue is damaged, and glucosinolates come into contact with plant myrosinase, a β-thioglucosidase. In intact tissue, this enzyme is stored separate from glucosinolates (1, 2). Myrosinase removes the β-glucose moiety from glucosinolates, leading to the formation of an unstable intermediate, and, finally, to a variety of toxic breakdown products (Fig. 1A). These compounds have diverse biological activity ranging from feeding deterrence for generalist insects to oviposition stimulation for specialists (4). Nevertheless, these breakdown products may also be toxic to crucifer specialists including diamondback moth (DBM) (8). The host range of this world-wide pest includes crops such as cabbage, broccoli, cauliflower, collards, rapeseed, mustard, and Chinese cabbage. Pest management is difficult, as DBM rapidly developed resistance to many synthetic insecticides (9, 10) and to microbial Bacillus thuringiensis toxin sprays (11, 12). It is also costly, exceeding U.S. $1 billion per year (13) in addition to the costs of crop loss. To use crucifers as host plants, DBM circumvents the chemical defense posed by the mustard oil bomb. We show that a sulfatase activity enables DBM to desulfate glucosinolates, which renders them invisible to myrosinase, allowing DBM to avoid the formation of toxic glucosinolate breakdown products. Our results have important ecological and evolutionary implications and suggest a new target for DBM pest management.
Fig 1.
Reactions catalyzed by plant myrosinase and diamondback moth GSS. (A) Myrosinase removes glucose from glucosinolates (Top), leading to the formation of toxic hydrolysis products (isothiocyanates, nitriles, thiocyanates; Bottom Left). (B) GSS forms desulfo-glucosinolates (Bottom Right).
Materials and Methods
Plant Material.
Arabidopsis thaliana seeds from the Col-0 (stock no. N1092) and Cvi-0 (stock no. N1096) ecotypes were kindly provided by the Nottingham Arabidopsis Stock Center, Nasturtium officinale and Brassica oleracea oleracea var. Rosella were obtained from Saatgut (Quedlinburg, Germany). A. thaliana and B. oleracea were grown under 11.5-h light/12.5-h dark cycles at 23°C in 5 × 5 or 8 × 8 cm2 pots on a 1:3 vermiculite/standard soil (Einheitserdenwerk, Fröndenberg, Germany) mix; N. officinale was grown partly submerged in 15 × 15 × 25 cm3 plastic boxes filled with standard soil and water under otherwise identical conditions.
Animals.
A Plutella xylostella stock culture (G88 colony) was kindly provided by A. M Shelton from Cornell University (Ithaca, NY). Larvae were reared on a wheat germ based artificial diet at 27°C and 16-h light/8-h dark cycles. Trichoplusia ni eggs were obtained from Entopath (Easton, PA), Spodoptera exigua pupae were obtained from Bayer (Wuppertal, Germany). Larvae were reared on DBM diet at room temperature. Pieris rapae was collected from a cabbage field near Jena, Germany; larvae were reared on brussels sprouts (Brassica oleraceae oleraceae). Before HPLC analysis of feces, all insects were reared on crucifers. Helix pomatia was collected near the Max Planck Institute for Chemical Ecology, and fed with Lactuca sativa or turnip. After feces analyses, snails were released at their original location.
Glucosinolate Sulfatase (GSS) Assay.
Fourth-instar larval tissue was ground in 100 mM Tris, pH 7.5. Debris was removed by centrifugation. Protein concentration in the supernatant was determined by using standard methods. One microgram of protein in 50 μl of 100 mM Tris, pH 7.5, was incubated with 50 μl of an aequeous solution of 5 mM glucosinolate for 3 min at room temperature. Reaction was stopped with 500 μl of methanol. After purification with 10 mg DEAE sephadex A-25 and centrifugation, supernatant was dried and dissolved in 400 μl of deionized water. Forty microliters were HPLC analyzed as described below.
Glucosinolate Extraction and HPLC Analysis.
Leaf samples (50–70 mg) were frozen in liquid nitrogen, freeze-dried, and ground to a fine powder. Glucosinolates were extracted with 500 μl methanol for 6 h, and twice with 500 μl of 60% methanol for 1 h. After centrifugation to remove debris, the supernatant was dried and dissolved in 400 μl of deionized water. After overnight incubation with 50 μg sulfatase from H. pomatia, crude extract, type H-1 (Sigma), purification with 10 mg DEAE sephadex A-25 and centrifugation, 40 μl from the supernatant were separated on a water (Solvent A)–acetonitrile (Solvent B) gradient at a flow rate of 1 ml/min and at 20°C by HPLC (Agilent HP1100 Series) fitted with a Lichrocart 250–4 RP18e 5-μm column. The 25 min run consisted of 1.5% B (1 min), 1.5–5.0% B (5 min), 5.0–7.0% B (2 min), 7.0–25.0% B (7 min), 25.0–92.0% B (3 min), and a 6 min hold at 92.0% B, followed by 92.0–1.5% B (1 min). Eluent was monitored by diode array detection between 190 and 360 nm (2 nm interval). Desulfo-glucosinolates were identified by retention time and UV spectra as compared with those of purified standards and quantified by A229 nm. Response factors determined from pure desulfo-glucosinolates were used to calculate molar concentrations of individual glucosinolates. Analysis of feces followed a similar protocol, but omitting the sulfatase step.
Synthesis of cDNA.
Total RNA was isolated with Trizol (Life Technologies) according to the manufacturer's recommendations. First-strand cDNA was synthesized from 1 μg total RNA each by using the Omniscript RT-kit (Qiagen, Hilden, Germany). Rapid amplification of cDNA ends (RACEs) followed a protocol modified from (14). Gene specific primers were Sulfi-R1: 5′-ATGGCNGAYGAYATGGGNTGGGA-3′ (3′-RACE) and Sulfi-F1: 5′-CCNACRTGCCAYTTNCCNACCAT-3′ (5′-RACE), respectively.
Reaction products were gel purified (QiaQuick; Qiagen) and cloned into pCRII–TOPO (Invitrogen). Inserts were sequenced on an ABI 3700 DNA sequencer with Big Dye Terminators (Applied Biosystems). Assembly and comparison of DNA sequence data were done by using dnastar (DNAstar, Madison, WI).
Heterologous Expression in Escherichia coli.
GSS cDNA encoding the entire ORF was amplified by using the primers SulForFull: 5′-ATATGGATCCATGCATTCAACCAACATGGCGATTC-3′ and SulRevFullStop: 5′-ATATGAATTCTTACAACTTTCACGGCGAACTGC-3′. The PCR product was gel purified, digested with BamHI and EcoRI, and cloned directionally into an appropriately cut pET28a(+) vector (Novagen). Correct insert sequence and reading frame was verified by sequencing. GSS constructs were expressed in E. coli One Shot BL21 Star (DE3) (Invitrogen) following the manufacturer′s recommendations.
Heterologous Expression in Spodoptera frugiperda Sf9 Cells.
GSS cDNA was amplified by PCR using the primers SulfFEcoRV: 5′-ATATGATATCAACATGGCGATTCTGCATCAAGC-3′ and SulfRNotI: 5′-ATATGCGGCCGCTTACAACTTTCACGGCGAACTGC-3′. The PCR product was digested with EcoRV and NotI, and ligated into pIZT/V5–His insect cell expression vector (Invitrogen). The GSS vector construct was propagated in E. coli TOP 10 cells. Correct orientation and sequence was confirmed by sequencing. For initiation of a Sf9 cell culture, a frozen cell stock was thawed at 37°C and added to 25 cm2 flasks with 4 ml of complete TNM-FH medium (Invitrogen) supplemented with 10% heat-inactivated FBS. The flasks were transferred to a 27°C incubator and cells were allowed to attach for 45 min. Afterwards, medium was exchanged and cells were controlled daily until a confluent monolayer had formed. Sf9 cells were maintained at 27°C in TNM-FH medium supplemented with 10% heat-inactivated FBS and 10 μg/ml of gentamycin. Transfection of Sf9 cells with either the GSS construct or a control vector construct (pIZT/V5-His/CAT) was performed by using Insectin-Plus liposomes as recommended by the manufacturer (Invitrogen). Transfected cells were grown in TNM-FH medium for 2, 3, and 5 days, respectively. Expression was driven by the Qrygia pseudotsugata multicapsid nucleopolyhedrosis virus immediate-early 2 (OpIE2) promoter. Supernatant from transfected Sf9 cells (1 ml per 25 cm2 flask) was collected at various times starting 24 h after transfection and pooled. Three milliliters of pooled supernatant from either the GSS or the control constructs was concentrated and buffer was exchanged by using Ultrafree-15 centrifugation units (Millipore). From 200 μl concentrated supernatant in 100 mM Tris⋅HCl (pH 7.5), 10 μl were used for enzyme assays as described above; the CAT construct was used as a negative control.
Reverse Transcription–PCR.
Ten fourth-instar larvae each were dissected in ice-cold Tris⋅HCl, pH 8, and separated into heads, body (without gut), silk glands, guts, and hindguts with Malpighian tubules. The guts were further cut into ≈2-mm long pieces. From these tissues, as well as from eggs, first, second, third, and fourth larval instars, prepupae, and adults, total RNA was isolated and reverse-transcribed into cDNA as described. GSS-specific primer pairs (Dsulf-F1: 5′-GTGGTGCTCCTCGGCGCGGC-3′/Dsulf-R2: 5′-AGCGTCCTGTAGGTACTGCGAGA-3′) were multiplexed with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers (GAPDH-F: 5′-CAGTGCCGATGCACCTATGTTC-3′/GAPDH-R: 5′-AAGTTGTCGTTGAGGGAGATGCC-3′). Reaction mixes contained 0.2 μl first strand cDNA, 2.5 μl 10× reaction buffer (Qiagen), 0.5 μl dNTPs (10 mM each), 1 unit Taq polymerase (Qiagen), 10 pmol of each GSS-specific primer, and 20 pmol of each GAPDH primer in a total volume of 25 μl. Cycling conditions were 94°C for 5 min followed by 25 cycles of 94°C, 58°C and 72°C for 30 s each, followed by 72°C for 7 min on a GeneAmp PCR System 9700 thermal cycler (Applied Biosystems).
Protein Electrophoresis and Blotting.
Native protein electrophoresis was performed on nondenaturing 8% (vol/vol) acrylamid/bisacrylamid (37.5:1) minigels using 0.3% (wt/vol) imidazol/0.8% (wt/vol) Hepes (pH 7.4) as a running buffer. SDS/PAGE were performed on 10% (vol/vol) acrylamid/bisacrylamid (37.5:1) resolving gels combined with 6% (vol/vol) stacking gels and using 0.1% (wt/vol) SDS/0.3% (wt/vol) Tris/1.4% (wt/vol) glycine (pH 8.3) as a running buffer. Gels were stained with Bio-Safe Coomassie G250 and/or Bio-Rad Silver Stain (Bio-Rad). Blotting onto Optitran BA-S85 membranes (Schleicher & Schüll) was carried out according the manufacturer's recommendations. Immunoblot detection using a polyclonal rabbit antibody (Eurogentec, Brussels) raised against heterologously expressed and nickel-agarose column purified GSS (Xpress System Protein Purification; Invitrogen) followed the instructions from the ECL Western blotting analysis system (Amersham Pharmacia).
Results and Discussion
Diamondback Moth Encodes a GSS Gene.
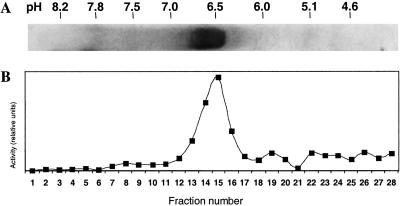
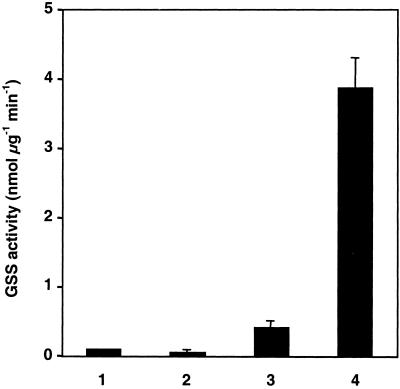
A common procedure to analyze glucosinolates involves sulfatase from H. pomatia. Glucosinolates are bound to anion exchange columns and released as desulfo-glucosinolates by treating with sulfatase (15). HPLC analysis of water/methanol extracted feces from DBM larvae fed on a variety of A. thaliana ecotypes revealed the presence of desulfo-glucosinolates. Also, in an expressed sequence tag collection from DBM larvae several, partly overlapping clones with strong similarity to sulfatase genes from various species were detected. This finding suggested that DBM contains a sulfatase activity that may desulfate glucosinolates, a GSS (Fig. 1B). To test this hypothesis, crude protein extracts from fourth-instar larval guts were separated on native PAGE gels. GSS activity was monitored with sinigrin, and correlated with a single band on a Coomassie brilliant blue stained gel. This band was excised from the native gels and subjected to SDS/PAGE, where it also migrated as a single band, with a molecular mass of approximately 66 kDa. These results were confirmed by isoelectric focusing (Fig. 2). Moreover, the band comigrating with GSS activity was excised from native gels, purified, desalted and concentrated with Centricon spin columns (Millipore) and, after lyophilization, subjected to matrix-assisted laser desorption ionization mass fingerprinting and electrospray ionization MS/MS (Wita Proteomics, Teltow, Germany). Only a single polypeptide was identified that exhibited the fingerprint pattern expected from GSS. Further analyses located GSS activity in the gut content and, in lesser amounts, in gut tissue but not in the remaining parts of the larval body (Fig. 3), indicating that the enzyme is secreted into the gut lumen.
Fig 2.
Isoelectric focusing and activity of purified GSS. (A) Isoelectric focusing of 10 μg natively purified DBM-GSS at pH 10 to 3, double stained with Bio-Safe Coomassie G250 and silver (Bio-Rad). (B) A further lane from the isoelectric focusing gel, loaded with 10 μg natively purified GSS, was divided vertically into 28 parts of 2 mm length. Gel pieces were suspended in 100 μl of 100 mM Tris⋅HCl, pH 7.5, and 30 μl each were tested for GSS activity. Vertical axis shows relative amounts of sinigrin converted to its desulfo-form, as measured by HPLC. Maximum GSS activity was detected in fraction 15.
Fig 3.
GSS activity in DBM larval tissue. Lane 1, Tris⋅HCl control; lane 2, body without gut; lane 3, gut tissue (without hindgut and Malpighian tubules); lane 4, gut content. Enzyme assays were performed as described. From each sample, 1 μg total protein was used. Guts were rinsed with Tris⋅HCl to remove remaining content.
Full-length cDNA sequence revealed an ORF of 1,641 bp, equivalent to a polypeptide of 547 aa (Fig. 4) or 62 kDa. psort ii (16) predicted a 19-aa signal peptide for extracellular targeting. Database searches revealed approximately 30% identity and 40–50% sequence similarity to arylsulfatases from various human and animal origins, but no particular affiliation to any database entry. Several consensus sequences were obtained resulting from allelic variation in our DBM laboratory strain (GenBank accession nos. AJ410304, AJ410305, AJ410306). Southern blots were compatible with the presence of a single GSS gene.
Fig 4.
(Upper) Exon–intron structure of the DBM GSS gene. Triangles mark binding sites for primers used in reverse transcription–PCR. (Lower) Amino acid sequence of DBM GSS. prosite sulfatase signatures are shaded gray. A predicted (16) signal sequence for extracellular targeting is printed in italics and underlined. Conserved amino acids of the catalytic center are underlined and printed extra bold. A highly conserved cystein residue posttranslationally converted into Cα-formylglycin in eukaryotic sulfatases (17) is shaded black.
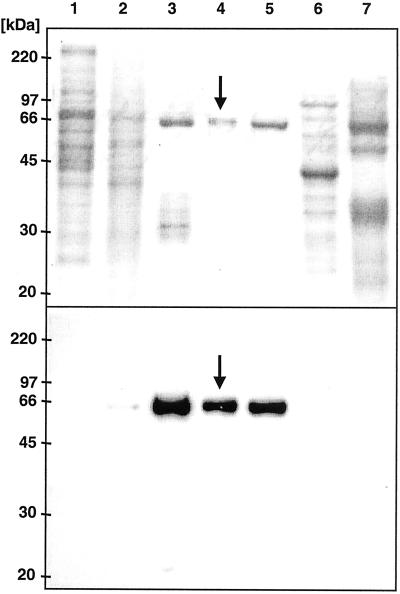
GSS heterologously expressed in E. coli did not show sulfatase activity, suggesting a lack of posttranslational modifications necessary to activate sulfatases (17). A polyclonal rabbit antibody was raised against nickel-agarose column purified protein extracts from E. coli heterologously expressing GSS. In immunoblots including a second, horseradish peroxidase coupled anti-rabbit antibody, the first antibody strongly reacted with a single band of ≈66 kDa in the gut content sample (Fig. 5). Most importantly, the antibody detected the natively purified polypeptide that comigrated with the GSS activity, demonstrating that we successfully cloned the GSS gene from DBM. Further evidence was obtained by functional expression in transformed insect Sf9 cells, which yielded low GSS activity, most likely reflecting a low transformation rate, but clearly distinct from experiments with control constructs that lacked GSS activity.
Fig 5.
Immunoblots localize DBM GSS to larval guts. (Upper) SDS/PAGE protein gel. Molecular mass is indicated on the left. Lane 1, 5 μg body without gut; lane 2, 5 μg gut tissue; lane 3, 3 μg gut content; lane 4, 0.1 μg native PAGE-purified DBM GSS (indicated by an arrow); lane 5, 1 μg Nickel-agarose purified GSS, heterologously expressed by E. coli; lane 6, 5 μg E. coli total protein with control vector without GSS insert; lane 7, 5 μg Helix pomatia sulfatase, crude extract, type H-1 (Sigma). (Lower) Immunoblot detection with polyclonal GSS antibody including a second anti-rabbit antibody. For immunoblotting one-tenth of the protein amounts from Upper was loaded, except for lane 4, where one-half was used. Adjustment of protein amounts was carried out to optimize staining.
GSS Expression Is Under Tissue-Specific and Developmental Control.
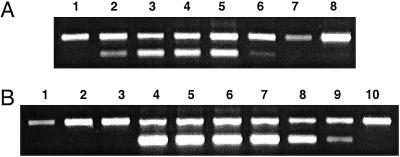
GSS expression was analyzed with RT PCR (Fig. 6). Specific primer pairs were multiplexed with GAPDH primers to control for variation in cDNA amounts synthesized from different RNA samples. GSS transcripts were detected in gut tissue but not the remaining body parts, which corresponds with the localization of GSS activity (Fig. 3). Furthermore, transcripts were detected in all four larval instars but not in DBM eggs, pupae, or adults, indicating that GSS expression is both under tissue-specific and developmental control.
Fig 6.
Reverse transcription–PCR analysis of GSS-specific mRNA. Upper bands indicate GAPDH control (549 bp), lower bands GSS (324 bp). (A) Developmental transcript analysis of GSS in DBM. Lane 1, eggs; lanes 2 to 5, first, second, third, and fourth larval instars; lane 6, prepupae; lane 7, pupae; lane 8, adults. (B) Spatial transcript analysis of GSS in fourth-instar larvae. Lane 1, head; lane 2, body (without gut); lane 3, silk glands; lanes 4 to 9, gut parts, from anterior to posterior; lane 10, hindgut with Malpighian tubules.
GSS Acts on Distinct Glucosinolate Classes.
Previous experiments in our laboratory showed no correlation between glucosinolate profiles of 38 A. thaliana ecotypes and resistance to DBM feeding. These ecotypes represented a collection of samples with very different mixtures of glucosinolate types and amounts (5), suggesting that GSS is able to act on different glucosinolate classes. Indeed, commercially available sinigrin (2-propenyl glucosinolate, Sigma) and glucotropaeolin (benzyl glucosinolate, Calbiochem) and purified intact (18) glucobrassicin (3-indoylmethyl glucosinolate), which have distinct side chain structures, were all rapidly converted to desulfo-glucosinolates both by crude DBM gut extract or by natively purified GSS (Table 1).
Table 1.
GSS enzymatic reaction constants for aliphatic, aromatic, and indolic glucosinolates
| GS (common name) | GS (scientific name) | GS type | n | Km, mM | vmax, μmol⋅mg−1⋅min−1 |
|---|---|---|---|---|---|
| Sinigrin | 2-Propenyl GS | Aliphatic | 3 | 1.61 ± 0.10 | 168.7 ± 7.6 |
| Glucotropaeolin | Benzyl GS | Aromatic | 3 | 1.40 ± 0.13 | 174.9 ± 7.6 |
| Glucobrassicin | 3-Indoylmethyl GS | Indolic | 2 | 2.33 ± 0.30 | 66.6 ± 7.6 |
GSS amount was quantified photometrically. 0.2 μg natively purified protein were used in each assay. Reactions were carried out for 3 min in 75 mM Tris⋅HCl, pH 7.5, at 24°C, then 400 μl methanol were added to stop the reaction, and desulfo-glucosinolates were quantified by HPLC. Sinigrin and glucotropaeolin were tested in concentrations from 0.1 mM to 25 mM; for glucobrassicin the maximum concentration was 10 mM, because of limitations in the amount of purified substrate. GS, glucosinolate. Given are mean values ± SE; n indicates the number of experimental replicates.
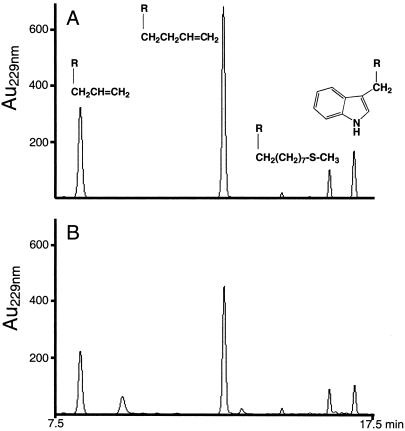
Moreover, GSS activity was tested in vivo on two A. thaliana ecotypes (Col-0 and Cvi-0) and N. officinale, containing high amounts of aliphatic, indolyl, or aromatic glucosinolates. Leaves from these plants were divided along their major veins, and veins were discarded. Glucosinolates were extracted directly from one half, including a Helix sulfatase treatment (15). The other halves were fed to fourth-instar DBM larvae reared on artificial diet (19), and feces were HPLC analyzed without prior Helix sulfatase treatment. In each case, HPLC profiles from corresponding leaf and feces samples revealed identical desulfo-glucosinolate peaks (Fig. 7). Feces samples contained approximately 25–40% less desulfo-glucosinolates than the direct extracted samples, indicating some glucosinolate degradation (Table 2). As DBM larvae do not feed continually (i.e., periods of heavy feeding alternate with pauses of various length), this difference may be explained by myrosinase activity in damaged leaf areas degrading glucosinolates pre ingestion.
Fig 7.
HPLC chromatograms of glucosinolate profiles. (A) A. thaliana Cvi-0 leaf halves. (B) Feces of fourth-instar larvae feeding on the other halves. Extraction, separation and identification were performed as described. Shown are signals from 7.5–17.5 min. Secondary structures indicate peaks caused, from left to right, by sinigrin (2-propenyl GS), gluconapin (3-butenyl GS), glucohirsutin (8-methylsulfinyloctyl GS), and glucobrassicin (3-indolylmethyl GS).
Table 2.
Quantification of major glucosinolates in leaves and desulfo-glucosinolates in larval feces
| Plant source | GS (common name) | GS (scientific name) | n | Leaves, μmol⋅g−1 dry weight | Feces, μmol⋅g−1 dry weight | Ratio, % |
|---|---|---|---|---|---|---|
| A. thaliana (Cvi-0) | Sinigrin | 2-Propenyl GS | 8 | 26.6 ± 2.3 | 19.0 ± 1.4 | 72 ± 4.2 |
| A. thaliana (Cvi-0) | Gluconapin | 3-Butenyl GS | 8 | 35.6 ± 3.0 | 26.7 ± 2.3 | 76 ± 4.5 |
| A. thaliana (Col-0) | Glucoraphanin | 4-Methylsulfinylbutyl GS | 10 | 11.4 ± 1.1 | 7.7 ± 0.7 | 68 ± 3.2 |
| A. thaliana (Col-0) | Glucobrassicin | 3-Indolylmethyl GS | 7 | 4.8 ± 0.2 | 2.8 ± 0.2 | 59 ± 2.8 |
| N. officinale | Gluconasturtiin | 2-Phenylethyl GS | 15 | 64.8 ± 2.7 | 41.1 ± 2.5 | 63 ± 2.2 |
Listed are plant sources and predominant glucosinolates (GS), number of experimental replicates (n), glucosinolate content in leaf halves, and desulfo-glucosinolate content in feces from fourth-instar larvae fed on corresponding leaf halves, and recovery ratios in feces vs. leaves. Given are mean values ± SE.
The gut content of a fourth-instar DBM larva reared on artificial diet contains ≈5–7 μg protein, equal to a GSS activity of 20–28 nmol/min (Fig. 2). Total glucosinolate amount of 100 mg fresh leaves is approximately 150 nmol for A. thaliana Col-0, and 700 nmol for Cvi-0 or N. officinale. Therefore, one larva could desulfate the glucosinolates present in 100 mg fresh leaves in 30 min. According to our observations, one DBM fourth-instar larva devours approximately 100 mg of fresh leaves in 50 h. Thus, actual GSS activity exceeds the minimal activity necessary to convert total glucosinolates in the plant meal by 100-fold.
In the insect gut, GSS competes with myrosinase for glucosinolate substrates. As myrosinases are not able to use desulfo-glucosinolates as a substrate (20) and sulfate competitively inhibits myrosinase (21), GSS could act in two ways, directly by removing the myrosinase's substrate, glucosinolates, and indirectly by reducing its activity by means of the released sulfate.
Ecological and Evolutionary Implications.
A simplified, but fundamental, concept of chemical ecology assumes that antagonistic chemical interactions between plants and herbivorous insects coevolve in a stepwise process: an advance in plant defenses exerts selective pressure on the insect's ability to overcome these defenses, and vice versa (22, 23). Diversification of defenses in a plant species is, thus, the outcome of a historical process, which may result from a “coevolutionary arms race” between the host and its pests. However, DBM can evade host plant diversification of glucosinolate structures, which are rapidly degraded by GSS. Also, variation in total glucosinolate content among crucifers does not affect herbivory by DBM (8, 24, 25), because of the excess of GSS activity in the larval gut (above). Consequently, in DBM herbivory, competition between myrosinase and GSS activity may determine the efficacy of glucosinolate defenses. Indeed, elevated myrosinase activity leads to reduced DBM damage in Brassica (8).
DBM feeds on many species of Brassicaceae. The wide substrate range of GSS is a prerequisite for the survival of newly hatched larvae on different cruciferous host plants, as female DBM are equally attracted by different glucosinolates when searching for oviposition sites (26). GSS may have been recruited from other metabolic pathways as DBM evolved specialization to crucifer host plants. Tight developmental and tissue-specific control ensures that GSS expression is limited to the stage at which DBM is exposed to glucosinolates and to the organ where these compounds are released from the ingested plant meal, consistent with an advanced stage in the evolution of a novel metabolic pathway (27). Nevertheless, there are further, yet unknown, ways to cope with glucosinolate–myrosinase based defenses. Crucifers are also hosts for Spodoptera exigua, Trichoplusia ni and Pieris rapae (28). However, in larvae from these species, we did not detect a GSS activity.
Nonetheless, H. pomatia, a generalist herbivore, also possesses a sulfatase that can desulfate glucosinolates. A crude fraction containing this activity is widely used for biochemical applications, e.g., for glucosinolate analysis. The gene encoding this activity has not yet been cloned (29). Therefore, it is difficult to determine whether the DBM and the Helix enzymes share a common origin. However, our polyclonal DBM GSS antibody did not cross-hybridize with the commercially available Helix sulfatase (Fig. 3). Moreover, desulfo-glucosinolates were detectable in feces from H. pomatia only when animals were fed with filter paper soaked with 5 mM sinigrin solution, but not when snails were allowed to feed on crucifers. Thus, Helix sulfatase appears unable to compete with plant myrosinase for the glucosinolate substrates, suggesting that the Helix glucosinolate sulfatase activity does not play a major role under natural conditions.
A New Target for Pest Management?
As glucosinolate breakdown products are toxic to DBM (8), impairing GSS activity would render the larvae susceptible to the host plant‘s defenses. Therefore, GSS could serve as a new target for DBM pest management. This may be achieved by developing GSS inhibitors. So far, limitations in the amount of pure GSS extractable from DBM guts have precluded this approach. However, this will be possible after optimization of GSS expression in Sf9 cells. Alternately, DBM mutants lacking GSS activity could be raised and released to mate with wild-type DBM.
Acknowledgments
We thank Domenika Schnabelrauch and Antje Figuth for sequencing. This work was supported by the Max Planck Society.
Abbreviations
GSS, glucosinolate sulfatase
DBM, diamondback moth
GAPDH, glyceraldehyde-3-phosphate dehydrogenase
References
- 1.Matile P. H. (1980) Biochem. Physiol. Pflanz. 175, 722-731. [Google Scholar]
- 2.Bones A. M. & Rossiter, J. T. (1996) Physiol. Plant. 97, 194-208. [Google Scholar]
- 3.Halkier B. A. (1999) in Naturally Occurring Glycosides: Chemistry, Distribution and Biological Properties, ed. Ikan, R. (Wiley, New York), pp. 193–223.
- 4.Rask L., Andreasson, E., Ekbom, B., Eriksson, S., Pontoppidan, B. & Meijer, J. (2000) Plant Mol. Biol. 42, 93-113. [PubMed] [Google Scholar]
- 5.Kliebenstein D. J., Kroymann, J., Brown, P., Figuth, A., Pedersen, D., Gershenzon, J. & Mitchell-Olds, T. (2001) Plant Phys. 126, 811-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kroymann J., Textor, S., Tokuhisa, J. G., Falk, K. L., Bertram, S., Gershenzon, J. & Mitchell-Olds, T. (2001) Plant Phys. 127, 1077-1088. [PMC free article] [PubMed] [Google Scholar]
- 7.Raybould A. F. & Moyes, C. L. (2001) Heredity 87, 383-391. [DOI] [PubMed] [Google Scholar]
- 8.Li Q., Eigenbrode, S. D., Stringam, G. R. & Thiagarajah, M. R. (2000) J. Chem. Ecol. 26, 2401-2419. [Google Scholar]
- 9.Furlong M. J. & Wright, D. J. (1994) Pesticide Sci. 42, 315-326. [Google Scholar]
- 10.Iqbal M. & Wright, D. J. (1997) B. Entomol. Res. 87, 481-486. [Google Scholar]
- 11.Tabashnik B. E., Liu, Y. B., Malvar, T., Heckel, D. G., Masson, L., Ballester, V., Granero, F., Mensua, J. L. & Ferre, J. (1997) Proc. Natl. Acad. Sci. USA 94, 12780-12785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heckel D. G., Grahan, L. J., Liu, Y. B. & Tabashnik, B. E. (1999) Proc. Natl. Acad. Sci. USA 96, 8373-8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Talekar N. S. & Shelton, A. M. (1993) Annu. Rev. Entomol. 38, 275-301. [Google Scholar]
- 14.Frohman M. A., Dush, M. K. & Martin, G. R. (1988) Proc. Natl. Acad. Sci. USA 85, 8998-9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thies W. (1979) Naturwissenschaften 66, 364-365., 1979. [Google Scholar]
- 16.Nakai K. & Horton, P. (1999) Trends Biochem. Sci. 24, 34-36. [DOI] [PubMed] [Google Scholar]
- 17.Dierks T., Lecca, M. R., Schlotterhose, P., Schmidt, B. & von Figura, K. (1999) EMBO J. 18, 2084-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kliebenstein D. J., Lambrix, V. M., Reichelt, M., Gershenzon, J. & Mitchell-Olds, T. (2001) Plant Cell 13, 681-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shelton A. M., Cooley, R. J., Kroening, M. K., Wilsey, W. T. & Eigenbrode, S. D. (1991) J. Entomol. Sci. 26, 17-26. [Google Scholar]
- 20.Ettlinger M. G., Dateo, G. P., Jr., Harrison, B. E., Mabry, T. J. & Thompson, C. P. (1961) Proc. Natl. Acad. Sci. USA 47, 1875-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shikita M., Fahey, J. W., Golden, T. R., Holtzclaw, W. D. & Talalay, P. (1999) Biochem. J. 341, 725-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ehrlich P. R. & Raven, P. H. (1964) Evolution (Lawrence, Kans.) 18, 586-608. [Google Scholar]
- 23.Berenbaum M. R. & Zangerl, A. R. (1998) Proc. Natl. Acad. Sci. USA 95, 13743-13748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bodnaryk R. P. (1997) Can. J. Plant Sci. 77, 283-287. [Google Scholar]
- 25.Kliebenstein D., Pedersen, D., Barker, B. & Mitchell-Olds, T. (2002) Genetics 161, 325-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reed D. W., Pivnick, K. A. & Underhill, E. W. (1989) Entomol. Exp. Appl. 53, 277-286. [Google Scholar]
- 27.Copley S. D. (2000) Trends Biochem. Sci. 25, 261-265. [DOI] [PubMed] [Google Scholar]
- 28.Agrawal A. A. (2000) OIKOS 89, 493-500. [Google Scholar]
- 29.Wittstock U., Fischer, M., Svendsen, I. & Halkier, B. A. (2000) IUBMB Life 49, 71-76. [DOI] [PubMed] [Google Scholar]