Abstract
Biological invasions are drastically altering natural habitats and threatening biodiversity on both local and global levels. In one of the United States' worst invasions, Eurasian Tamarix plant species have spread rapidly to dominate over 600,000 riparian and wetland hectares. The largest Tamarix invasion consists of Tamarix chinensis and Tamarix ramosissima, two morphologically similar species. To clarify the identity, origins, and population structuring of this invasion, we analyzed DNA sequence data from an intron of a nuclear gene, phosphoenolpyruvate carboxylase (PepC). This intron proved to be highly variable at the population level, and the 269 native and invasive specimens yielded 58 haplotypes, from which we constructed a gene genealogy. Only four of these haplotypes were common to both the U.S. and Eurasia. Surprisingly, we found that the most common plant in this U.S. invasion is a hybrid combination of two species-specific genotypes that were geographically isolated in their native Eurasian range. Less extensive hybrids exist in the invasion, involving combinations of T. ramosissima and T. chinensis with Tamarix parviflora and Tamarix gallica. The presence of potentially novel hybrids in the U.S. illustrates how importation of exotics can alter population structures of species and contribute to invasions.
Invasion of nonnative species into natural areas ranks second behind only habitat destruction as the largest ecological disaster worldwide (1). Of the 972 plants and animals listed by the United States' Endangered Species Act, approximately 400 are at risk primarily because of competition with and predation by nonnative species (2). In addition to this ecological damage, the economic impact of invasive species on agriculture, forestry, and public health in the United States is estimated to total $123 billion per year (3). For these reasons, the control of selected invasives is becoming an integral part of ecosystem stewardship.
The ability of invasive plants to compete and proliferate can be caused by intrinsic factors such as physiological or reproductive capacities often associated with weedy species (4), and to extrinsic factors such as loss of competitors, herbivores, or pathogens upon introduction. An additional influence may be the creation of novel genotypes. Introduction of a species into a new region can bring into contact closely related species or genotypes that had previously been geographically isolated. This contact creates new opportunities for hybridization events, which may produce genotypes with high fitness in the invaded habitat. These novel genotypes may also be able to deter pathogens and herbivores as well as, or even better than, parental genotypes. Through molecular investigation, the number of plant invasions found to contain hybrids is increasing, and the role of this evolutionary process may have been undervalued in previous studies of invasion (5).
Several species of the genus Tamarix L. (common name: saltcedar or tamarisk, family Tamaricaceae) are, as a group, considered one of the worst plant invasions in the U.S., exceeded only by purple loosestrife (Lythrum salicaria) (2). Tamarix is an Old World genus of approximately 54 shrub and tree species, found in salty, dry, or riparian habitats. The plants can outcross, self pollinate, and also propagate clonally from woody fragments (6). The small white to pink clustered flowers are pollinated by numerous species of insects, and possibly by the wind (7), and the small seeds have hairs on one end that enable long-distance wind dispersal.
Eight to twelve species were brought to the U.S. from southern Europe or Asia in the 1800's to be used for shade and erosion control (8), and a subset of these species has taken over more than 600,000 riparian and wetland hectares (6). This invasion is expanding by 18,000 hectares per year (9) through many western U.S. natural areas, including major river systems and national parks. During its importation to the U.S., Tamarix escaped nearly all of its biological enemies (10) and has proven difficult to control on a large scale by either manual or chemical methods. Intrinsic characteristics associated with weedy plants, such as vegetative reproduction and high seed output (6), may also have aided their success. Here we report an additional and unexpected factor contributing to the invasion: widespread hybridization of two species-specific genotypes, an event that was undetected in the native Eurasian range.
Researchers at the United States Department of Agriculture (Agricultural Research Service) are currently searching for and testing candidate biological control insects as an alternative means of suppressing this invasion (10, 11). Initial biological control tests of the saltcedar leaf beetle (Diorhabda elongata) show differential survival on what appears to be a single species of Tamarix collected from different regions of the U.S., grown in common garden plots (11). Insects can be species-specific, and in some cases, host-specificity can reach to the level of the plant genotype (e.g., refs. 12 and 13). The reduced survival of insects on certain plants may have been caused by variation in the physiological condition of the plants (11), but genotypic differences in the plants could have also influenced the results.
Many species of Tamarix are widespread in Eurasia (14), and it is unlikely that all of the genotypes of any one species were imported to the U.S. Unfortunately, historical records do not reveal precise origins or genetic information concerning the introductions (15). The control agents being tested may not have evolved with certain invasive genotypes, and thus result in ineffective or suboptimal control. For these reasons, biological control researchers would like to know how many genotypes are represented in the U.S. invasion, and to what scale we can pinpoint their Eurasian origins.
A previous study of the morphology and phylogenetic placement of invasive Tamarix suggests that there are at least four invasive entities in the U.S., including Tamarix aphylla, Tamarix parviflora, morphologically similar Tamarix canariensis and Tamarix gallica, and the largest invasion consisting of morphologically similar Tamarix chinensis and Tamarix ramosissima (16). These last two species are by far the most common in the U.S., and they are the focus of this investigation.
T. chinensis is native to China, Mongolia, and Japan, whereas T. ramosissima is widespread from eastern Turkey to Korea (14). The ranges of these two species putatively overlap for approximately 4,200 km across China to Korea. Invasive T. ramosissima and T. chinensis plants are noted from many areas of the western U.S., and T. ramosissima extends into southern Canada and northern Mexico. In the latest revision of the genus, T. chinensis and T. ramosissima are placed in different taxonomic sections and are morphologically distinguished by a few microscopic floral characters (especially where the filament is inserted into the nectary disk) and edaphic affinities (T. ramosissima is halophilous, whereas T. chinensis is not) (14). Alternatively, some botanists claim that their morphology is similar (e.g., ref. 17), and that it is difficult to recognize the two taxa as different species, let alone assign them to different sections of the genus. Our previous molecular work confirms the close relationship between the species (16), but we show support for their distinction below.
To determine plant invasion identities and origins, analyses traditionally match genotypes from native and invasive populations using data such as allozymes, RAPDs (random amplified polymorphic DNA), and AFLPs (amplified fragment length polymorphism) (e.g., refs. 18–20). The advantage of these methods is in the ability to process large amounts of population samples, and the good potential for finding genetic variation. DNA sequence data can also determine identities and origins of invasives and, in addition, gene genealogies constructed from these ordered data will demonstrate the mutational relationships of existing alleles (21). The amount of invasive plant population studies using DNA sequence data has been few because of the lack of markers that show population level resolution (22), and the relatively time consuming and costly processing of samples. Additionally, sequence markers only represent a small portion of the genome, and genotypes must be inferred from these small regions. However, as bulk sequencing becomes more efficient and high-resolution markers slowly become available, researchers are beginning to use sequence data to study the geographic distribution of invasive plant genotypes (e.g., ref. 23), and the future of this area of invasive plant research looks promising.
Materials and Methods
A total of 269 vouchered DNA samples of Tamarix were collected from the western U.S. (155 plants) and Eurasian native populations of T. chinensis and T. ramosissima (114 plants), with 1–8 individuals per population. J.F.G. made many of the collections, especially from the U.S., Iran, Republic of Georgia, Turkmenistan, and Kazakstan. In these cases, 6–8 plants were sampled from a population. Various collectors generously provided all other samples. As discrimination between T. chinensis, T. ramosissima, and their hybrids is not possible without examination of the nectary disk under a dissection microscope, plants were selected in the field without regard to their taxonomic status, as long as they were one of the two species of concern. The identities of all plants were later determined with Baum's morphological descriptions and keys (14). Phylogenetically informative intraspecific markers are scarce for most plants, especially noncrop species. Many published markers were evaluated, and either did not amplify, were multicopy, or did not contain sufficient variation. We used conserved coding region (exon) sequences from closely related crop plants in the order Caryophyllales (e.g., spinach, carnation) to design primer pairs and amplify potentially variable noncoding regions (introns) of several nuclear genes not previously used in population analyses. Of these, the fourth intron of the phosphoenolpyruvate carboxylase gene (PepC) proved highly variable within T. ramosissima.
Genomic DNA was isolated from silica-dried fresh leaf tissue. The fourth PepC intron region was amplified by PCR using primer pair PPCL1 (forward) (5′-GTCCCTAAGTTTCTGCGTCG-3′) and PPCL2 (reverse) (5′-CTTCAGGTGTTACTCTTGGG-3′) with the following cycling conditions: 95°C (2 min); 30 cycles of 95°C (1 min), 50°C (1 min), 72°C (2 min); and then 32°C (5 min). A 50-μl reaction was performed for each individual, and PCR products were purified by agarose gel electrophoresis followed by QIAquick Gel Extraction kit (Qiagen, Valencia, CA). Purified templates were sequenced in two directions by using the dideoxy chain termination method with ABI prism Dye Terminator Cycle Sequencing Ready Reaction kit with AmpliTaq DNA polymerase (Perkin–Elmer). Specimens were electrophoresed in an ABI 373A automated sequencer following manufacturer's instructions (Applied Biosystems). For heterozygous specimens, identities of the two haplotypes (alleles) were inferred using “haplotype subtraction” (24). To check our haplotype inferences, we cloned PepC alleles from a variety of heterozygous individuals using pGEM-T Vector System II (Promega) and then sequenced these using the protocol above. To determine the number of copies of the fourth intron of PepC in the genome, DNA was digested according to the manufacturer's (New England Biolabs) recommendations using enzymes EcoRI and HindIII. Southern blots were prepared and hybridized as described in Jeddeloh et al. (25), using a radiolabeled fragment encompassing the fourth PepC intron.
Results and Discussion
The DNA region we sequenced was approximately 900 bases in length and contained 144 variable sites in the species investigated. Fifty-seven percent of all of the individuals assayed were heterozygous, and alleles of the 30 heterozygous individuals that we cloned matched the inferred haplotypes in all cases. A Southern analysis of the PepC intron yielded only a single size fragment for both T. ramosissima and T. canariensis, evidence that this gene is either single or low copy.
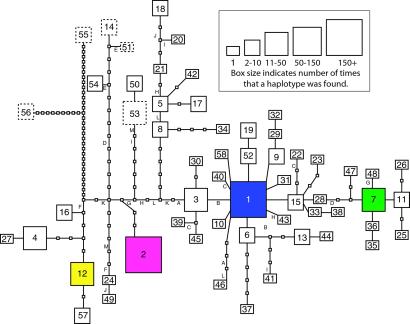
A total of 58 haplotypes were found among the 269 individuals (a total of 538 alleles) (Appendix 1, which is published as supporting information on the PNAS web site, www.pnas.org). All populations except one in China (T. chinensis) had more than one haplotype represented, and some had up to 11 different haplotypes in six plants (Appendix 2, which is published as supporting information on the PNAS web site). We used these haplotypes to manually construct a maximum parsimony gene genealogy or minimum spanning network (Fig. 1), which represents the mutational relationships among the haplotypes. Thirteen (9.0%) of the 144 mutations were homoplasious (i.e., they appeared in more than one place on the gene genealogy).
Fig 1.
The PepC fourth intron gene genealogy. Boxes with numbers (1 through 58) represent haplotypes (alleles) recovered. The small empty boxes represent intermediate haplotypes not recovered in this analysis. Lines separating the haplotype boxes, no matter what their length, represent a single point mutation or insertion/deletion event. Haplotypes in solid line boxes are T. ramosissima or T. chinensis, and those in colored boxes are found in both Eurasia and U.S. Haplotypes in dashed boxes are known from species other than T. ramosissima or T. chinensis. Letters next to lines represent homoplasious mutations.
A decline in variability is expected after founder events or genetic bottlenecks. The U.S. T. chinensis and T. ramosissima invasion had lower overall genetic diversity than the native range, with 12 of the total 58 haplotypes (20.6%) compared with the 50 haplotypes found in Eurasia (86.2%). Four haplotypes (6.9%) were found in both Eurasia and the U.S. (see Tables 1 and 2), and eight haplotypes were found only in the U.S. (the basis for this is discussed below). We found a total of 80 genotypic combinations, with 56 (70%) of these exclusive to Eurasia, 20 (25%) exclusive to the U.S., and only 4 (5.0%) common to both areas.
Table 1.
T. ramosissima and T. chinensis haplotypes common to both native and invasive areas
| Haplotype | Number in Eurasia | % of total Eurasian alleles | Number in U.S. | % of total U.S. alleles |
|---|---|---|---|---|
| 1 | 62 | 27.2 | 121 | 39.0 |
| 2 | 58 | 25.4 | 114 | 36.7 |
| 7 | 7 | 3.1 | 4 | 1.3 |
| 12 | 2 | 0.9 | 32 | 10.3 |
Table 2.
T. ramosissima and T. chinensis genotypes common to both native and invasive areas
| Genotype | Number in Eurasia | % of Eurasian specimens | Number in U.S. | % of U.S. specimens |
|---|---|---|---|---|
| 1/1 | 11 | 9.6 | 32 | 20.6 |
| 1/2 | 0 | 0.0 | 33 | 21.3 |
| 1/7 | 2 | 1.6 | 2 | 0.6 |
| 1/12 | 0 | 0.0 | 14 | 9.0 |
| 2/2 | 28 | 24.6 | 30 | 19.3 |
| 2/7 | 0 | 0.0 | 1 | 0.3 |
| 2/12 | 0 | 0.0 | 5 | 1.6 |
| 7/12 | 0 | 0.0 | 1 | 0.3 |
| 12/12 | 1 | 0.9 | 5 | 3.2 |
The most common haplotypes were 1 and 2, which differed by 9 mutations (see Fig. 1). Haplotype 1 accounted for 27.2% of the alleles in Eurasia and 39.0% of those in the U.S. Haplotype 2 accounted for 25.4% of the alleles in Eurasia and 36.7% of those in the U.S. The next most common haplotypes were 12 (10.3% of the alleles in the U.S.) and 53 (5.5% of the alleles in the U.S.). The remaining haplotypes each represented less than 5% of the alleles in either the U.S. or Eurasia.
Levels of heterozygosity varied intrinsically between the two species in their native range. We detected 54 different genotypic combinations for 81 T. ramosissima specimens in Asia, and 76.3% of these were heterozygous. Tamarix chinensis, on the other hand, had two genotypes in 29 Asian specimens, and only 6.6% of these were heterozygous. These results are consistent with observations made in the 19th century that T. chinensis in China is extensively cultivated and rarely found in the wild (26).
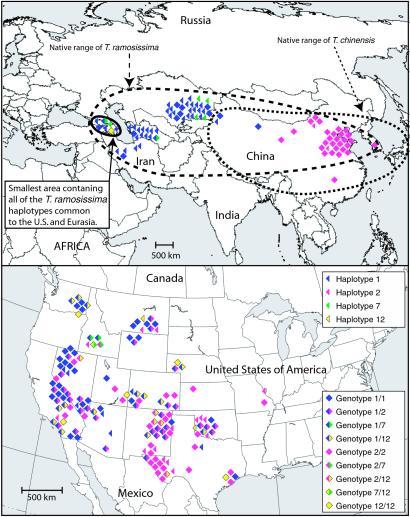
In the native Eurasian range, by far the most common genotypic combinations were 1/1 and 2/2, which belong to T. ramosissima and T. chinensis, respectively (see Tables 1 and 2). In contrast, within the U.S. invasion these genotypes were the second and third most common. The most common genotype in the U.S. was 1/2, a morphologically cryptic hybrid of T. ramosissima and T. chinensis which we did not detect in Eurasia. Even though both species are putatively found all across China (14), we found the 1/1 genotype exclusively west of central China and the 2/2 genotype exclusively east of central China (see Fig. 2). In Asia there are no known physical barriers between the two species except their putative edaphic affinities (14), and in the U.S. we found the T. chinensis (2/2) and T. ramosissima (1/1) genotypes growing together in 5 populations, and once they were growing within 2 m of each other on disturbed homogenous soil.
Fig 2.
Approximate distribution of T. chinensis and T. ramosissima haplotypes and genotypes common to Eurasia and the U.S. in their native range (above) and in the United States (below). Locations of specimens are spread out on map to avoid overlapping. Bold circle indicates smallest Eurasian area that contains all T. ramosissima haplotypes common to the U.S. and Eurasia. Dashed circles indicate native range of species sensu Baum (14).
Haplotype 12 was the third most common in the U.S., found throughout the invasion, and it differs from haplotype 1 by 14 mutations. We found it only once in Eurasia in a homozygous plant (genotype 12/12) in Azerbaijan. The 2/12 genotype, found only in the U.S., may be another novel hybrid (although perhaps a very rare one) given the disjunct native range of the haplotypes (China and Azerbaijan, respectively). Haplotype 7 was the only other T. ramosissima or T. chinensis haplotype common to the U.S. and Eurasia. It differs from haplotype 1 by 5 mutations, and was found in only one population in Idaho; in Eurasia it was found in the Republic of Georgia, Turkmenistan, and Kazakstan.
The smallest native region that contains all of the detected T. ramosissima haplotypes common to Eurasia and the U.S. (1, 7, and 12) consists of the Republic of Georgia and Azerbaijan (see Fig. 2). Designating the native range of invasive genotypes has practical applications for biological control agent searches involving pest species with a widespread native distribution, but would require more extensive sampling than we have provided in this study.
Three haplotypes, 51, 53, and 55, detected in U.S. T. ramosissima or T. chinensis specimens, are known haplotypes from other invasive species (T. gallica and T. parviflora) that are native to the Mediterranean region west of the range of T. ramosissima. They are genetically relatively distant from haplotype 1 (see Fig. 2) and were found (but only rarely) in the U.S. in combination with the most common haplotypes, 1 and 2, which belong to T. ramosissima and T. chinensis respectively. Given the allopatry of these species' native ranges, it is possible that invasive genotypic combinations of T. ramosissima or T. chinensis and the other invasive Tamarix species (e.g., 1/53, 1/55, 2/53) have also originated recently because of human movement and cultivation. Whether the hybridization first occurred in the U.S. or Europe is unknown, as T. ramosissima is used horticulturally in Europe. Another four haplotypes found in the U.S., 50, 54, 56, and 57, are also relatively genetically distant from haplotype 1 of T. ramosissima. These haplotypes may have also originated from other species because we did not detect them in the native range of T. ramosissima and T. chinensis.
Tamarix are still sold in some areas of the U.S., and there has been some concern that the horticultural specimens are contributing to the invasion. We found haplotype 56 in the commonly sold T. ramosissima “Pink Cascade” and in two invasive specimens, indicating that contribution of genomic material from this cultivar to the invasion may be rare, but does exist. What the genetic contribution of this cultivar to the invasion will be in the future is unpredictable. The last haplotype detected exclusively in the U.S., 52, differs from haplotype 1 by only one mutation. Being genetically similar to T. ramosissima, it is interesting that we did not find it in the native range of T. ramosissima in Eurasia.
Conclusions
These data, taken in total, indicate little if any hybridization among T. ramosissima and T. chinensis in their native range, even though their ranges putatively overlap. Sampling was not thorough in western and central China, and the 1/2 genotype may certainly occur in these areas, but given the overlap of the two species' ranges, we were surprised that we did not find T. ramosissima haplotypes in the eastern half of China, where they putatively exist. In contrast, we found extensive hybridization among two of the invasive Tamarix species within the U.S. The 1/2 genotype, representing a T. chinensis × T. ramosissima hybrid, is the most common plant found in the invasion, ranging from Oklahoma to Washington to California. Less extensive hybrids exist in the invasion, involving combinations of T. ramosissima and T. chinensis with T. parviflora and T. gallica. The abundance of cryptic hybrids helps explain why identification of species in the U.S. using morphology has been, and will continue to be, problematic.
What do these results mean for the biological control of Tamarix? An effective and safe control agent should have high host specificity, the result of a shared evolutionary history. The United States' hybrid Tamarix genotypes may be as little as 200 years old (15), and thus have essentially no shared evolutionary history with any genotype-specific predators or diseases. Proposed Tamarix control agents such as the Asian leaf beetle (Diorhabda elongata deserticola) and the tamarisk leaf weevil (Coniatus tamarisci) have not yet been tested on the 1/2 hybrid (J.F.G., unpublished data). The presence of a successful novel hybrid in the U.S. invasion may potentially confound biological control results, depending on the control agent's level of host specificity. Moreover, the results reported here allow us to circumscribe a native area that contains all detected T. ramosissima haplotypes common to both the U.S. and Eurasia, information that may help focus future biological control agent searches.
Analyses of highly variable nuclear DNA sequence data allow us to understand the diversity and distribution, and mutational relationships of plants in both their native and introduced ranges, and in this study we have been able to track some of the history and origins of the recent T. chinensis and T. ramosissima invasion. Multiple introductions of exotic Tamarix have brought together formerly isolated genotypes, allowing hybridization to occur and alter the genetic structure of Tamarix in the U.S. The widespread presence of hybrid Tamarix in its introduced range serves as an additional warning for how continued accidental or intentional importation of numerous plant species can unexpectedly alter the genotypic composition of naturalized populations and potentially contribute to the ecological devastation caused by exotic species invasion.
Supplementary Material
Acknowledgments
We thank members of the Schaal lab, C. D'Antonio, W. J. Leverich, and A. Snow for helpful comments. R. Crocker, C. J. DeLoach, J. Friedman, F. Ghahremani-nejad, E. Konno, A. Miller, I. D. Mityaev, I. Nee, Y. Qin-Er, J. Schulte, P. Shafroth, J. Shippen, J. Taylor, J. L. Tracy, and G. Yatskievych generously donated specimens. This work was supported by U.S. Department of Agriculture National Research Initiative Competitive Grants Program, Cooperative State Research, Education, and Extension Service Grant 2000-00836 (to B.A.S. and J.F.G.), National Geographic Society Committee for Research and Exploration Grant 6663-99 (to J.F.G.), the Mellon Foundation support of Missouri Botanical Garden graduate students, and an Environmental Protection Agency Science To Achieve Results (STAR) graduate fellowship (to J.F.G.).
References
- 1.Wilson E. O., (1997) Strangers in Paradise (Island, Washington, DC).
- 2.Stein B. A. & Flack, S. R., (1996) America's Least Wanted: Alien Species Invasions of U.S. Ecosystems (The Nature Conservancy, Arlington, VA).
- 3.Pimentel D., Lach, L., Zuniga, R. & Morrison, D. (2000) Bioscience 50, 53-65. [Google Scholar]
- 4.Baker H. G. (1965) in The Genetics of Colonizing Species, eds. Baker, H. G. & Stebbins, G. L. (Academic, New York).
- 5.Ellstrand N. C. & Schierenbeck, K. A. (2000) Proc. Natl. Acad. Sci. USA 97, 7043-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brotherson J. D. & Field, D. (1987) Rangelands 3, 110-112. [Google Scholar]
- 7.Shmida A. (1991) Israel Land and Nature 16, 119-125. [Google Scholar]
- 8.Baum B. R. (1967) Baileya 15, 19-25. [Google Scholar]
- 9.DiTomaso J. M. (1998) Weed Technol. 12, 326-336. [Google Scholar]
- 10.DeLoach C. J., Carruthers, R. I., Lovich, J. E., Dudley, T. L. & Smith, S. D. (2000) in Proceedings of the X International Symposium on Biological Control of Weeds, ed. Spencer, N. (Montana State Univ., Bozeman), pp. 819–873.
- 11.DeLoach C. J. & Tracy, J. L., (1997) The Effects of Biological Control of Saltcedar (Tamarix ramosissima) on Endangered Species: Biological Assessment Draft (U.S. Department of Agriculture—Agricultural Research Service, Temple, TX).
- 12.Rank N. E. (1991) in Natural History of Eastern California and High-Altitude Research, eds. Hall, C. A., Doyle-Johnes, V. & Widawski, B. (White Mountain Research Station, Univ. of California, Bishop), pp. 161–181.
- 13.Schoonhoven L., Jermy, T. & van Loon, J., (1998) Insect–Plant Biology (Chapman and Hall, New York).
- 14.Baum B. R., (1978) The Genus Tamarix (Israel Acad. Sci. Hum., Jerusalem).
- 15.Horton J. S., (1964) Notes on the Introduction of Deciduous Tamarix (U.S. Forest Service, Fort Collins, CO).
- 16.Gaskin, J. F. & Schaal, B. A. (2002) Syst. Bot. 28, in press.
- 17.Crins W. J. (1989) J. Arnold Arbor. 70, 403-425. [Google Scholar]
- 18.Novak S. J. & Mack, R. N. (2001) Bioscience 51, 114-122. [Google Scholar]
- 19.Garcia P., Vences, F. J., Pérez de la Vega, M. & Allard, R. W. (1989) Genetics 122, 687-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh R. & Jain, S. (1971) Theoret. Appl. Genet. 41, 79-84. [DOI] [PubMed] [Google Scholar]
- 21.Avise J. C. (1989) Evolution (Lawrence, Kans.) 43, 1192-1208. [DOI] [PubMed] [Google Scholar]
- 22.Schaal B. A., Hayworth, D. A., Olsen, K. M., Rauscher, J. T. & Smith, W. A. (1998) Mol. Ecol. 7, 465-474. [Google Scholar]
- 23.Saltonstall K. (2002) Proc. Natl. Acad. Sci. USA 99, 2445-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clark A. G. (1990) Mol. Biol. Evol. 7, 111-112. [DOI] [PubMed] [Google Scholar]
- 25.Jeddeloh J. A., Bender, J. & Richards, E. J. (1998) Genes Dev. 12, 1714-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helmsley A. L. S. (1888) J. Linn. Soc. Bot. 23, 346. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.