Abstract
Genetic heterozygosity is thought to enhance resistance of hosts to infectious diseases, but few tests of this idea exist. In particular, heterozygosity at the MHC, the highly polymorphic loci that control immunological recognition of pathogens, is suspected to confer a selective advantage by enhancing resistance to infectious diseases (the “heterozygote advantage” hypothesis). To test this hypothesis, we released mice into large population enclosures and challenged them with multiple strains of Salmonella and one of Listeria. We found that during Salmonella infections with three avirulent strains, MHC heterozygotes had greater survival and weight than homozygotes (unlike sham controls), and they were more likely to clear chronic Salmonella infection than homozygotes. In laboratory experiments, we found that MHC heterozygosity enhanced the clearance of multiple-strain Salmonella infections. Yet, contrary to what is widely assumed, the benefits of heterozygosity were due to resistance being dominant rather than overdominant, i.e., heterozygotes were more resistant than the average of parental homozygotes, but they were not more resistant than both. The fact that MHC heterozygotes were more resistant to infection and had higher fitness than homozygotes provides a functional explanation for MHC-disassortative mating preferences.
The highly polymorphic genes of the MHC play a central role in the immune recognition of pathogens and parasites. These genes encode MHC class I and II molecules that present peptide antigens to T cells, initiating immune recognition of foreign antigens (1). Heterozygosity at MHC loci may enhance resistance to infectious diseases by increasing the diversity of antigens presented to T cells (2), and by generating a diverse T cell repertoire (3). The evidence for the heterozygote advantage hypothesis, however, is mixed and equivocal (4, 5). MHC heterozygosity is associated with resistance to some infectious diseases, including more effective clearance of hepatitis B virus (6), lower viral loads among carriers of human T cell lymphotropic virus (HTLV-I) (7), and prolonged survival of HIV-infected individuals (8). Yet, these findings are correlations that may be due to genetic heterozygosity at background loci, and moreover, just as many population studies report no association between MHC heterozygosity and resistance to infectious diseases (9–11).
Experimental studies with laboratory mice (Mus domesticus) provide evidence that MHC heterozygosity increases immune resistance (at least for 9 of 16 of the pathogens tested) (5), but some problems remain unresolved. First, host resistance is often tested by using immunological responses and pathogen clearance assays (8 of 16 studies), which do not necessarily indicate how a host survives or copes with infection. For example, one study found that heterozygous mice had higher T cell responses to a viral infection than homozygotes, and yet heterozygotes were least likely to survive the infection (2). (The authors suggested that high immune responsiveness of heterozygotes might provide an advantage during natural infections, but this possibility was never tested.) Second, most studies only tested 2–3 genotypes, limiting the conclusions that can be made. Third, laboratory studies usually use congenic strains of mice to control non-MHC effects, but crossing inbred lines to produce heterozygotes may create an incidental heterozygote advantage because of heterosis, i.e., a spurious effect resulting from masking recessive deleterious mutations that may accumulate in inbred lines (12). Fourth, experimental studies have only tested single strains of pathogens or parasites, whereas the evidence for heterozygote advantage in human populations has been with genetically polymorphic infections (6, 8). Thus, the critical tests of the MHC-heterozygote advantage hypothesis have not been performed.
Another problem is that when heterozygotes show an advantage in population studies (6–8), it is generally interpreted as “heterozygote superiority” or “overdominance” (i.e., heterozygotes are assumed to have higher fitness than both parental homozygotes). Yet, heterozygotes may have a selective advantage over homozygotes on average if resistance is merely dominant (i.e., if heterozygotes inherit the resistance of the most resistant parental homozygote); and resistance is generally dominant, at least in tests with single pathogens (5). Therefore, for clarity, we will use “heterozygote advantage” in the broad sense when heterozygotes have a fitness advantage over homozygotes on average (like population studies), which may be due to dominance or overdominance, and only use heterozygote superiority (or overdominance) in the narrow sense when heterozygotes have higher fitness than both parental homozygotes. This distinction is meant to clarify whether an observed heterozygote advantage is due to resistance being overdominant, or merely dominant.
To determine whether heterozygosity at MHC loci provides a selective advantage against multiple-strain infections, we bred congenic mice carrying five haplotypes to produce homozygotes or heterozygotes. We controlled for the potential confounding effects of background mutations that could create heterosis by using F2 segregants (bred by intercrossing heterozygotes and MHC-genotyping the progeny). We released the mice into large population enclosures, infected them repeatedly during a 30-week period (simulated epidemics) with 1–5 strains of Salmonella enterica serovar Typhimurium and one strain of Listeria monocytogenes, and then monitored subsequent impacts on host fitness. To determine how MHC-heterozygosity influences the expression of genetic resistance, we infected mice and monitored pathogen clearance in the laboratory. Our results indicate that MHC-heterozygosity provides a selective advantage by increasing resistance to avirulent multiple-strain infections, although this pattern was due to resistance being dominant rather than overdominant.
Materials and Methods
Animals.
We used MHC-congenic strains of mice (C57BL/10SnJ-H2b, B10.D2-H2d, B10.M-H2f, B10.BR-H2k, B10.Q-H2q) obtained from The Jackson Laboratory. These strains carry five different MHC haplotypes, whose alleles have major genetic differences at all class I and II loci (13–15). Mice were bred and housed in standard colony conditions in a specific pathogen-free animal facility, either as pure lines or F2 segregants (i.e., homozygotes and heterozygotes were bred by crossing F1 crosses of congenic strains), and housed with same-sex littermates, 3–5 per cage. F2 segregants were MHC-genotyped by using PCR of microsatellite markers closely linked to the MHC (16).
Population Enclosures.
Enclosures were housed in a 320-m2 heated building designed for mouse populations (17), which were not sterile and had previously housed wild mice, so other pathogens and parasites were probably present. We released 260 mice into 10 population enclosures (24.5-m2), with approximately equal sex ratios and equal numbers of mice (26 on average) in each enclosure. The mice carried 14 different MHC genotypes and were homozygous or heterozygous for five MHC haplotypes (b, d, k, q, f). They varied in their ages (20 ± 0.6 weeks old) at the beginning of the experiment, but no age difference between homozygotes and heterozygotes existed. We infected mice in five enclosures, sham-infected mice in the other five, and monitored the health and survival of the mice 1–3 times per week.
Pathogens.
S. enterica serovar Typhimurium is an enteric mouse pathogen that becomes systemic by invading the intestinal mucosa and replicating intracellularly within host macrophages (18). Pathogen clearance and pathogenesis depend on the host genotype and Salmonella strain (19), and MHC genes influence the clearance of avirulent strains (20–24). We infected mice with the following Salmonella strains: aroA, a highly attenuated strain (25, 26); 628, an avirulent strain in C57 mice and clearance is MHC-dependent (20); LT2, an avirulent strain in secondary, but fatal during primary infections of C57 mice (27); and PMAC45 and PMAC51, avirulent heterologous strains carrying recombinant antigens from polio virus and hepatitis B virus, respectively (28). We used these recombinant strains because they express different epitopes, and thus ensure that the multiple-strain infections were antigenically diverse. L. monocytogenes (RF4738) is an intracellular pathogen that is avirulent in C57 mice, and although resistance is influenced by class I and II-dependent T cell responses (29), resistance is controlled by a monomorphic MHC locus (30).
Experimental Infections.
We cultured bacteria (stored as frozen stocks at −70°C) in 20 ml of heart–brain infusion at 37°C for 12 h while shaking. We diluted the overnight solution with sterile PBS to the desired concentration and verified the concentration of viable bacteria by quantitative plate counts. In the enclosure populations, we infected mice orally (25 μl solution), after withholding food and water for 4 h. In the laboratory experiments, we infected mice interperitoneally by using 0.2-ml injections per mouse. For single-strain infections in the laboratory, we infected 60 mice (equal numbers of bb, bq, qq genotypes) with Salmonella [strain 628, 4 × 103 colony-forming units (cfu) per mouse], either as a primary or secondary infection (20 mice of each genotype and equal sex ratio). To test the effects of MHC heterozygosity on secondary immunity, we first infected mice with the attenuated aroA strain (4 × 103 cfu per mouse) (or we sham-infected the control mice that at this time) and allowed four weeks for the bacteria to be cleared. For multiple-strain infections in the laboratory, we tested mice with approximately equal numbers of bb, bq, qq, dd, dq genotypes using pure strains, and then bb, bd, dd when testing F2 segregant mice. We infected mice with the aroA strain (103 cfu per mouse), and 4 weeks later, we infected them with either a single strain (628) or a mixture containing equal numbers of four Salmonella strains (aroA, 628, PMAC45, and PMAC51) (103 cfu per mouse for both single and mixed infections). We used selective media (Salmonella-Shigella and Rambach agar) to confirm that the mice were infected with Salmonella.
Pathogen Resistance.
To assay pathogen clearance, we killed infected mice 7–14 days after infection (always controlling the duration of infection among experimental groups), and dissected and homogenized spleens (1 ml of PBS) under sterile conditions. We performed serial dilutions of spleen homogenates, cultured 50 μl of each homogenate on selective agar plates, and incubated overnight (37°C). We determined the concentration of bacteria per spleen (“pathogen load”) by counting the number of colony forming units per milliliter of spleen homogenate on the plates (with the mean of two replicate plates per mouse). We also assayed resistance by monitoring weight loss, mortality, and signs of pathology (i.e., a disfigured hunched back associated with cachexia). In the enclosures, weight provided an index for the health of males (whereas the weight of females fluctuated mainly from pregnancy).
Statistical Analyses.
We used nonparametric Kaplan–Meier tests to analyze survivorship and Fisher's exact or Log-likelihood for goodness-of-fit tests. Before statistical tests, we checked the data for assumptions of normality and equality of variances, and transformed all nonnormal data (or used Welch's F tests when the variances were still unequal) or used the nonparametric Wilcoxon test. Tukey–Kramer tests were used for multiple comparisons. Data are presented as mean ± SEM, and we used two-tailed tests or directed tests (to minimize the number of mice) (31) where indicated (i.e., when our previous data already showed significance or a strong trend in the predicted direction).
Results and Discussion
Enclosure Experiment.
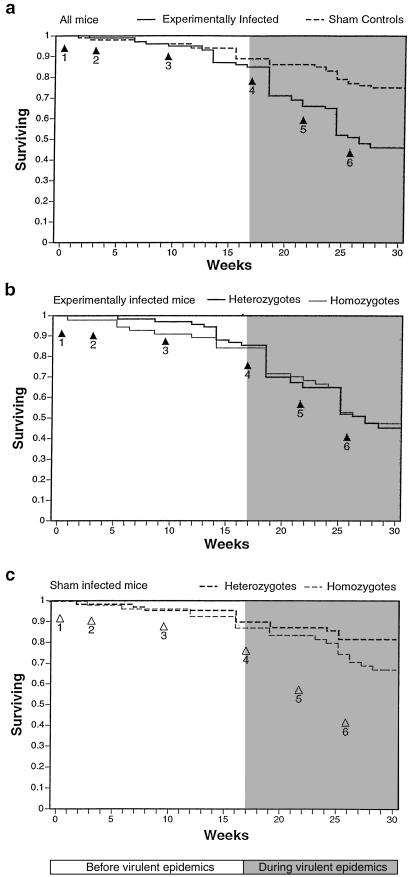
In the enclosure populations, the experimentally infected mice had lower survival than the sham-infected controls (P = 0.0001, n = 260), with mortality mainly occurring during the last three “virulent” infections after week 17 (Fig. 1a). Females had significantly higher survival rates than males in the sham-infected populations (P = 0.0004), whereas this pattern was reversed in the infected populations (P = 0.07). Overall, no survival advantage occurred for heterozygotes in the infected (P = 0.4) or sham-infected populations (P = 0.10). Yet, among the mice that died before the virulent infections (before week 17), MHC-heterozygous mice showed an advantage over homozygotes (P = 0.05; n = 20; Fig. 1b). In contrast, heterozygosity conferred no survival advantage among the mice that died during this time in the control populations (P = 0.8; n = 14; Fig. 1c), which implies that MHC heterozygosity prolonged survival among mice infected during the infections of avirulent Salmonella strains.
Fig 1.
Kaplan–Meier plots showing the survivorship of all of the mice (a), experimentally infected mice (b), and sham-control mice (c) in the population enclosures. Arrows indicate the timing of infections: (arrow 1) Salmonella aroA (2 × 106 cfu per mouse), (arrow 2) Salmonella 628 (106 cfu per mouse), (3) Salmonella LT2 (8 × 105 cfu per mouse), (arrow 4) Salmonella aroA, 628, LT2, PMAC45, and PMAC51 (total 106 cfu per mouse), and (arrows 5 and 6) two infections of Listeria (106 cfu per mouse).
Heterozygotes lost their survival advantage in the infected populations after the virulent infections (week 17; Fig. 1b), even though they showed an advantage in the control populations during this time (P = 0.05, n = 111; Fig. 1c). It is unclear why the virulent infections abolished the advantages of heterozygosity, although two of these were with Listeria, and resistance to this microbe is controlled by a monomorphic MHC locus (30). In the sham-infected populations, heterozygotes showed a survival advantage during the last 13 weeks of the experiment (when the mice were between 33 and 50 weeks of age). In laboratory conditions, C57 mice do not begin to show signs of age-related mortality until they are 57 weeks old. MHC effects do not become apparent until they are very old (between 85 and 132 weeks of age), and MHC heterozygotes show no advantage (32) or a disadvantage (33). Therefore, the reduced survival of homozygous mice in the control populations must have been caused by the conditions in the population enclosures, i.e., increased exposure to uncontrolled pathogens, social stress, or both. We found no evidence for Salmonella infection among the sham-infected mice, although some mice showed pathological symptoms of disease.
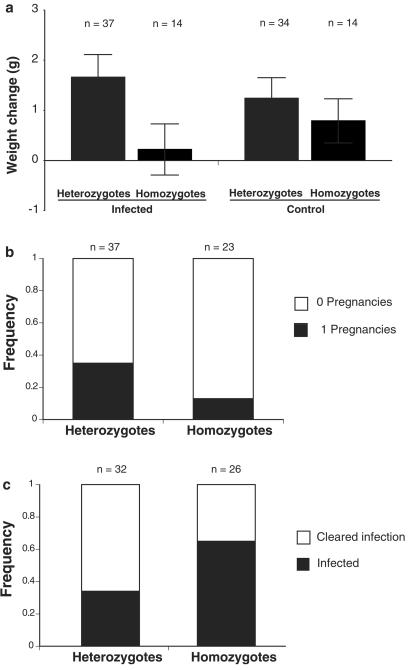
We also found that heterozygous mice were more healthy than homozygotes during the avirulent epidemics. In the infected populations, MHC-heterozygous males gained significantly more weight than homozygotes during the first two epidemics (P = 0.04, n = 69). This result was not necessarily due to our experimental infections, however. Uninfected, heterozygous mice in laboratory conditions do not show such a weight gain advantage (see below), but heterozygotes in the sham-control populations showed a significant weight-gain advantage during this time (P = 0.02, n = 60). This finding suggests that MHC heterozygosity may have provided an advantage against uncontrolled pathogens in the controls, but another (nonmutually exclusive) possibility is that the increased weights of heterozygotes are the result of genome-wide heterosis. We tested this possibility by examining the weights of F2 segregants, and found that among the F2 segregants, MHC heterozygotes gained significantly more weight than homozygotes in the infected populations, but not in the control populations (Fig. 2a). Thus, when we controlled for potential differences due to background genes, MHC heterozygotes still had a weight advantage, but only during experimental infections. We monitored the pregnancy status of females, and found that, in the infected populations, MHC heterozygotes were twice as likely to become pregnant (Fig. 2b) and they had more pregnancies than homozygotes (infected, P = 0.1, n = 60; controls, P = 0.35, n = 64), although these differences were not significant.
Fig 2.
Health and reproduction of mice in the population enclosures. (a) Changes in body weight of males before the virulent epidemics (F2 segregants; infected, P = 0.015, n = 51; controls, P = 0.4, n = 48). (b) Reproduction of females in infected populations (P = 0.076, n = 60) (controls, P = 0.80, n = 64, data not shown). (c) Mice at the end of the experiment from the experimental populations that were still infected or completely cleared the Salmonella infection (P = 0.034, n = 58).
Finally, we found that MHC-heterozygous mice were better able to resolve Salmonella infection than homozygotes. At the end of the 30-week experiment, we found that nearly half of the mice in the treatment populations were still infected with Salmonella. (This was unexpected, because under laboratory conditions these mice usually clear these strains within 1 month, and it had been 2 months since the last experimental Salmonella infection.) MHC heterozygotes were significantly more likely to have completely cleared infection than homozygotes (Fig. 2c). This result indicates that MHC heterozygosity enhances the ability of mice to clear chronic Salmonella infection under stressful, social conditions.
Laboratory Experiments.
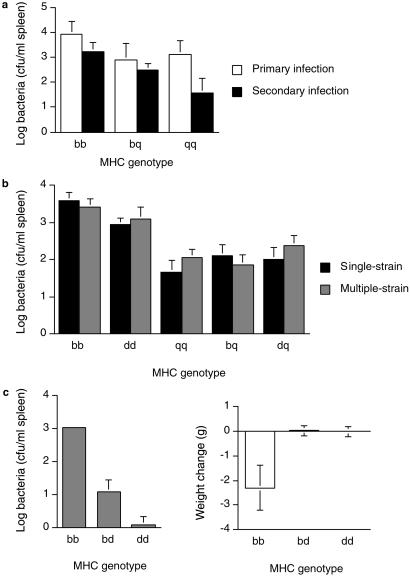
To determine whether the protective effects of MHC heterozygosity in the enclosures was caused by enhancing resistance against single- or multiple-strain infections, and whether resistance is dominant or overdominant, we conducted additional experiments in the laboratory. In the first experiment, we tested the resistance of mice against a single strain of Salmonella, either as a primary or as a secondary challenge (administered 3 weeks after a primary infection with an attenuated strain, as in the enclosures). Two weeks after infection, we determined the pathogen loads of the mice and found that MHC genotype (P = 0.03 n = 59) and infection type (primary or secondary, P = 0.007) had a significant effect on pathogen load (Fig. 3a). MHC-dependent resistance showed a dominant rather than an overdominant pattern of resistance. Heterozygotes had lower pathogen loads than homozygotes, especially in the secondary infections; however, these differences were not significant for primary or secondary challenges. Differences in resistance among homozygotes was magnified in the secondary infection (i.e., MHC genotype had a significant effect on pathogen load for the secondary, but not primary infection). This result is consistent with other studies (34) and studies showing that vaccines protect some but not all MHC genotypes (35–38), which may be because detectable levels of T cell responses against a Salmonella epitope are found during secondary and not primary infections (39). Therefore, we tested the resistance of MHC heterozygotes during secondary infections in subsequent experiments.
Fig 3.
Resistance of mice to Salmonella in the laboratory. (a) Pathogen loads of mice infected with a single strain (628), either as a primary (P = 0.46, n = 29) or secondary challenge (P = 0.04, n = 30). Heterozygotes were not significantly more resistant than homozygotes in the primary (P = 0.44, n = 29, power = 0.09, least significant number = 348) or the secondary infections (P = 0.11, n = 30, power = 0.11, least significant number = 87). (b) Pathogen loads of mice infected with secondary challenge of a single or multiple strains. On average, heterozygotes were significantly more resistant than homozygotes to the multiple-strain infections (P = 0.011), but not the single-strain infection (P = 0.077). (c) Resistance of F2 segregant mice after a secondary challenge of multiple Salmonella strains. MHC genotype had a significant influence on pathogen load (Pdir = 0.0001, n = 27) and weight (P = 0.01). On average, MHC-heterozygous mice had lower pathogen loads (Pdir = 0.04) and lost less weight (Pdir = 0.02) during infection than homozygotes.
In a second laboratory experiment, we tested the resistance of MHC-heterozygous and homozygous mice to multiple- or single-strain infections of Salmonella. After challenging mice with an attenuated aroA strain, we infected them with a secondary challenge of a mixture of four Salmonella strains, or as a control, we infected them with the same dosage of a single-strain (using a larger sample size of mice this time). MHC genotype had a significant effect on pathogen load for the single-strain (P = 0.0001, n = 67) and multiple-strain infections (P = 0.0001, n = 71) (Fig. 3b). Again, the mice showed a dominant rather than an overdominant pattern of resistance. (Heterozygotes were significantly more resistant than the susceptible parental genotypes, but they were never more resistant than both parental homozygotes.) On average, MHC heterozygotes had significantly lower bacterial loads than homozygotes in the multiple-strain, but not the single-strain infections.
In a third laboratory experiment, we tested the resistance of mice against multiple-strain infections of Salmonella by using F2 segregant mice (to rule out possible confounding effects of heterosis caused by background genes). We infected mice with four different Salmonella strains, and 1 week later, we found that MHC heterozygotes had significantly lower pathogen loads, and lost less weight during infection than homozygotes (Fig. 3c). Again, we found no significant evidence for heterozygote superiority. Heterozygotes were less likely to show pathological disease symptoms (a disfigured hunched back) than homozygotes, less likely than either parental homozygote (Pdir = 0.13, n = 26), although this trend was not significant. We found this overdominance pattern in a preliminary experiment (using pure lines of the same congenic mouse strains), although this trend is not statistically significant even when the results of the two experiments are pooled (Pdir = 0.09, n = 120). This experiment indicates that MHC heterozygotes are more resistant to secondary, multiple-strain infections than homozygotes, and it rules out the possibility that the benefits of heterozygosity are an artifact from crossing different congenic strains of mice (heterosis).
Summary and Implications.
Our results indicate that MHC heterozygosity confers a selective advantage against avirulent, multiple-strain Salmonella infections. In the population enclosures, MHC heterozygosity slightly enhanced the health and survival of mice during avirulent infections of different Salmonella strains, and enhanced the clearance of chronic infection. Heterozygotes showed a survival advantage in our control populations during the final weeks of the experiment, which may have been due to enhancing resistance to various uncontrolled pathogens in the enclosures. In the laboratory, MHC heterozygotes were more resistant to multiple-strain infections than homozygotes. This pattern was found in all of the genotypic combinations that we tested and could not be attributed to heterosis from masking background mutations that may have accumulated in these mice. Although MHC heterozygotes were more resistant than homozygotes on average, we found no significant evidence for heterozygote superiority. MHC heterozygotes were more resistant than homozygotes on average, but they were never significantly more resistant than the most resistant parental homozygote.
Our findings suggest that the associations between MHC heterozygosity and disease resistance in population studies (6–8) could be due to dominance rather than overdominance, contrary to what is generally assumed. Heterozygote superiority might emerge under special circumstances (4, 40, 41), but even if this would occur, overdominance seems to be insufficient to explain the evolution of MHC polymorphisms. Only one population genetic model concludes that overdominant selection can maintain MHC polymorphisms (42), but this result requires the unlikely assumption that heterozygous genotypes all have equal fitness. More realistic models indicate that overdominance can maintain only a few alleles (43, 44). On the other hand, if heterozygosity at MHC loci increases resistance to infectious diseases in the wild, then this finding could explain why MHC-heterozygous males have high reproductive success in macaques (45), increased antler development in white-tailed deer (46), and sexually attractive odors in stickleback fish (47). Furthermore, heterozygote advantage could explain the adaptive function of MHC-disassortative mating preferences in mice and humans (5), a behavior that is capable of driving the unusual diversity of MHC alleles (48).
Acknowledgments
We thank all of the students and animal-care staff who helped us with this research, especially T. Ellevold, J. Gale, B. Hopwood, J. Latham, E. McClelland, M. Perkins, S. Ross, M. Wilson, and S. Zala. We thank D. Ebert, B. Demarest, M. Milinski, and S. Zala for comments on the paper, and K. Bunny, E. Enioutina, and C. Hormaeche for helpful advice about Salmonella. Bacteria were kindly provided by C. Hormaeche (628), M. Hofnung (recombinant PMAC strains), J. Roth (aroA and LT2), and R. Daynes (Listeria). This research was supported by National Science Foundation Grant IBN9904609 and National Institutes of Health Grant GM39578.
Abbreviations
cfu, colony-forming unit
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Falk K., Rotzschke, O., Stevanovic, S., Jung, G. & Rammensee, H. G. (1991) Nature (London) 351, 290-296. [DOI] [PubMed] [Google Scholar]
- 2.Doherty P. C. & Zinkernagel, R. M. (1975) Nature (London) 256, 50-52. [DOI] [PubMed] [Google Scholar]
- 3.Dyall R., Messaoudi, I., Janetzki, S., Nikolic, Z. & Ugic, J. (2000) J. Immunol. 164, 1695-1698. [DOI] [PubMed] [Google Scholar]
- 4.Apanius V., Penn, D., Slev, P., Ruff, L. R. & Potts, W. K. (1997) Crit. Rev. Immunol. 17, 179-224. [DOI] [PubMed] [Google Scholar]
- 5.Penn D. J. (2002) Ethology 108, 1-21. [Google Scholar]
- 6.Thursz M. R., Thomas, H. C., Greenwood, B. M. & Hill, A. V. (1997) Nat. Genet. 17, 11-12. [DOI] [PubMed] [Google Scholar]
- 7.Jeffery K. J., Siddiqui, A. A., Bunce, M., Lloyd, A. L., Vine, A. M., Witkover, A. D., Izumo, S., Usuku, K., Welsh, K. I., Osame, M. & Bangham, C. R. (2000) J. Immunol. 165, 7278-7284. [DOI] [PubMed] [Google Scholar]
- 8.Carrington M., Nelson, G. W., Martin, M. P., Kissner, T., Vlahov, D., Goedert, J. J., Kaslow, R., Buchbinder, S., Hoots, K. & O'Brien, S. J. (1999) Science 283, 1748-52. [DOI] [PubMed] [Google Scholar]
- 9.Hill A. V., Allsopp, C. E., Kwiatkowski, D., Anstey, N. M., Twumasi, P., Rowe, P. A., Bennett, S., Brewster, D., McMichael, A. J. & Greenwood, B. M. (1991) Nature (London) 352, 595-600. [DOI] [PubMed] [Google Scholar]
- 10.Paterson S., Wilson, K. & Pemberton, J. M. (1998) Proc. Natl. Acad. Sci. USA 95, 3714-3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langefors A., Lohm, J., Grahn, M., Andersen, O. & von Schantz, T. (2001) Proc. R. Soc. London Ser. B 268, 479-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bailey D. W. (1982) Immunol. Today 3, 210-214. [DOI] [PubMed] [Google Scholar]
- 13.Pullen J. K., Horton, R. M., Cai, Z. L. & Pease, L. R. (1992) J. Immunol. 148, 953-967. [PubMed] [Google Scholar]
- 14.She J. X., Boehme, S. A., Wang, T. W., Bonhomme, F. & Wakeland, E. K. (1991) Proc. Natl. Acad. Sci. USA 88, 453-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edwards S. V., Chesnut, K., Satta, Y. & Wakeland, E. K. (1997) Genetics 146, 655-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saha B. K. & Cullen, S. E. (1986) J. Immunol. 136, 1112-1119. [PubMed] [Google Scholar]
- 17.Meagher S., Penn, D. J. & Potts, W. K. (2000) Proc. Natl. Acad. Sci. USA 97, 3324-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neidhardt F. C., (1996) Escherichia coli and Salmonella: Cellular and Molecular Biology (Am. Soc. Microbiol. Press, Washington, DC).
- 19.Benjamin W. H., Turnbough, C. L., Jr., Posey, B. S. & Briles, D. E. (1986) Infect. Immun. 51, 872-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hormaeche C. E., Harrington, K. A. & Joysey, H. S. (1985) J. Infect. Dis. 152, 1050-1056. [DOI] [PubMed] [Google Scholar]
- 21.Nauciel C., Ronco, E., Guenet, J. L. & Pla, M. (1988) Infect. Immun. 56, 2407-2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fayolle C., O'Callaghan, D., Martineau, P., Charbit, A., Clement, J. M., Hofnung, M. & Leclerc, C. (1994) Infect. Immun. 62, 4310-4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hess J., Ladel, C., Miko, D. & Kaufmann, S. H. (1996) J. Immunol. 156, 3321-3326. [PubMed] [Google Scholar]
- 24.Lo W. F., Ong, H., Metcalf, E. S. & Soloski, M. J. (1999) J. Immunol. 162, 5398-5406. [PubMed] [Google Scholar]
- 25.Hoiseth S. K. & Stocker, B. A. (1981) Nature (London) 291, 238-239. [DOI] [PubMed] [Google Scholar]
- 26.Hormaeche C. E., Khan, C. M. A., Mastroeni, P., Viia-real, B., Dougan, G., Roberts, M. & Chatfield, S. N. (1995) in Molecular and Clinical Aspects of Vaccine Development, eds. Ala'Aldeen, D. & Hormaeche, C. E. (Wiley, New York), pp. 119–153.
- 27.Xu H. R. & Hsu, H. S. (1992) J. Med. Microbiol. 36, 377-381. [DOI] [PubMed] [Google Scholar]
- 28.Charbit A., Newton, S. M., Klebba, P. E., Clément, J. M., Fayolle, C., Lo-Man, R., Leclerc, C. D. & Hofnung, M. (1997) Behring Inst. Mitt. 98, 135-142. [PubMed] [Google Scholar]
- 29.Ladel C. H., Flesch, I. E., Arnoldi, J. & Kaufmann, S. H. (1994) J. Immunol. 153, 3116-3122. [PubMed] [Google Scholar]
- 30.Seaman M. S., Wang, C. R. & Forman, J. (2000) J. Immunol. 165, 5192-5201. [DOI] [PubMed] [Google Scholar]
- 31.Rice W. R. & Gaines, S. D. (1994) Trends Ecol. Evol. 9, 235-237. [DOI] [PubMed] [Google Scholar]
- 32.Popp D. M. (1982) Mech. Ageing Dev. 18, 125-134. [DOI] [PubMed] [Google Scholar]
- 33.Salazar M., Leong, T., Tu, N., Gelman, R. S., Watson, A. L., Bronson, R., Iglesias, A., Mann, M., Good, R. A. & Yunis, E. J. (1995) Proc. Natl. Acad. Sci. USA 92, 3992-3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schito M. L., Chobotar, B. & Barta, J. R. (1998) Parasitol. Res. 84, 394-398. [DOI] [PubMed] [Google Scholar]
- 35.Sher A., Hieny, S. & James, S. (1984) Parasite Immunol. (Oxf.) 6, 319-328. [DOI] [PubMed] [Google Scholar]
- 36.Apt A. S., Avdienko, V. G., Nikonenko, B. V., Kramnik, I. B., Moroz, A. M. & Skamene, E. (1993) Clin. Exp. Immunol. 94, 322-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lo-Man R., Martineau, P., Deriaud, E., Newton, S. M., Jehanno, M., Clement, J. M., Fayolle, C., Hofnung, M. & Leclerc, C. D. (1996) Infect. Immun. 64, 4424-4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahlborg N., Ling, I. T., Holder, A. A. & Riley, E. M. (2000) Infect. Immun. 68, 2102-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McSorley S. J., Cookson, B. T. & Jenkins, M. K. (2000) J. Immunol. 164, 986-993. [DOI] [PubMed] [Google Scholar]
- 40.Penn D. & Potts, W. (1999) Am. Nat. 153, 145-164. [DOI] [PubMed] [Google Scholar]
- 41.Hughes A. L. & Nei, M. (1992) Genetics 132, 863-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takahata N. & Nei, M. (1990) Genetics 124, 967-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lewontin R. C., Ginzburg, L. R. & Tuljapurkar, S. (1978) Genetics 88, 149-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hedrick P. W. (1999) Genetica 104, 207-214. [DOI] [PubMed] [Google Scholar]
- 45.Sauermann U., Nurnberg, P., Bercovitch, F. B., Berard, J. D., Trefilov, A., Widdig, A., Kessler, M., Schmidtke, J. & Krawczak, M. (2001) Hum. Genet. 108, 249-254. [DOI] [PubMed] [Google Scholar]
- 46.Ditchkoff S. S., Lochmiller, R. L., Masters, R. E., Hoofer, S. R. & Van Den Bussche, R. A. (2001) Evolution (Lawrence, Kans.) 55, 616-625. [DOI] [PubMed] [Google Scholar]
- 47.Reusch T. B., Haberli, M. A., Aeschlimann, P. B. & Milinski, M. (2001) Nature (London) 414, 300-302. [DOI] [PubMed] [Google Scholar]
- 48.Hedrick P. W. (1992) Genetics 132, 575-581. [DOI] [PMC free article] [PubMed] [Google Scholar]