Abstract
Pathogens can be an important selective agent in plant evolution because they can severely reduce plant fitness and growth. However, the role of pathogen selection on plant evolution depends on the extent of genetic variation for resistance traits and their covariance with host fitness. Although it is usually assumed that resistance traits will covary with plant fitness, this assumption has not been tested rigorously in plant–pathogen interactions. Many plant species are tolerant to herbivores, decoupling the relationship between resistance and fitness. Tolerance to pathogens can reduce selection for resistance and alter the effect of pathogens on plant evolution. In this study, we measured three components of Arabidopsis thaliana resistance (pathogen growth, disease symptoms, and host fitness) to the bacteria Pseudomonas syringae and investigated their covariation to determine the relative importance of resistance and tolerance. We observed extensive quantitative variation in the severity of disease symptoms, the bacterial population size, and the effect of infection on host fitness among 19 accessions of A. thaliana infected with P. syringae. The severity of disease symptoms was strongly and positively correlated with bacterial population size. Although the average fitness of infected plants was smaller than noninfected plants, we found no correlation between the bacterial growth or symptoms expressed by different accessions of A. thaliana and their relative fitness after infection. These results indicate that the accessions studied vary in tolerance to P. syringae, reducing the strength of selection on resistance traits, and that symptoms and bacterial growth are not good predictors of host fitness.
In nature, plants are constantly challenged by disease-causing pathogens including viruses, bacteria, and fungi. Pathogen infection can severely reduce survivorship and reproduction of native plants, and in crops pathogen infection results in an estimated 12% loss of yield annually in the U.S. (1). Because pathogens are ubiquitous and strongly affect host plant fitness, pathogens are thought to be an important selective agent that shapes plant evolution. Previous studies suggest that pathogen-mediated selection affects a wide range of host plant traits including morphology, life history, mating system, and even community-level diversity (2, 3).
Pathogens can be a selective force on plant evolution only if three specific criteria are met (4). First, pathogen infection must affect host fitness; second, heritable variation in resistance traits must occur among individuals; and third, the heritable resistance trait must covary with the fitness of the plant host. The agriculture literature provides many examples of both yield reduction by pathogens, and heritable genetic variation for resistance among crop varieties (5–7). Evidence from natural populations is less extensive, but variation in resistance among populations has been documented (8–10), as well as fitness reduction due to pathogen infection (11, 12). However, the extent to which genetic traits that affect disease resistance covary with host fitness has not been established. Although, covariation between fitness and disease resistance seems obvious, it is possible that plants develop tolerance to pathogen infection, which decouples any association between disease resistance traits and host fitness. As a consequence, the effect of pathogen selection will be reduced when tolerance occurs.
The first challenge in studying the covariation between resistance and fitness is to accurately define and measure disease resistance. Resistance traits are broadly defined as host traits that reduce the extent of pathogen infection (4, 13). Thus, resistance traits are those that reduce host contact with pathogens, and those that reduce pathogen growth rate once infection has occurred. Empirically, resistance is measured in several different ways. Resistance in crop varieties is usually evaluated by their relative yield with and without pathogen infection (14–16). In contrast, genetic variation for disease resistance in natural populations is usually estimated by quantitative variation in visual symptoms (9, 12, 17, 18). These different estimates of plant resistance are equivalent if disease symptoms are a direct consequence of pathogen growth, and if fitness loss to pathogens is directly correlated with disease symptoms. However, the assumption that the amount of disease symptoms expressed are directly correlated with pathogen growth has not been specifically tested in plants [although previous studies that measured the two traits in A. thaliana suggest there is a correlation (e.g., ref. 19)]. Furthermore, studies on the interaction between plants and herbivores suggest that reduction in fitness does not necessarily correlate with the plant's ability to avoid herbivore damage (11, 20, 21). These studies show that plants vary not only in their resistance to herbivores, but also in their ability to tolerate the damage—i.e., plants that sustain higher infections or disease damage may not necessarily suffer a high reduction in fitness. Similarly, it is possible that resistance and tolerance to pathogens are controlled by different and uncorrelated traits. Some traits in plants may confer resistance by preventing host contact with pathogens or by reducing pathogen growth. The same or completely different host traits may increase host tolerance by diminishing the effect of infection on fitness. In plants where resistance and tolerance exists, yield comparisons between varieties will confound genetic variance for resistance and tolerance.
Tolerance to pathogen infection, defined as the host's ability to reduce the effect of infection on plant fitness (13), can have significant consequences to host and pathogen evolution. Disease resistance in the host population places strong selection on pathogens to evolve new genotypes that can avoid plant defenses. Thus, models of plant–pathogen interactions generate frequency-dependent selection that favor complex coevolutionary dynamics and maintain genetic polymorphism in both host and pathogen populations (22). In contrast, theoretical models show that if there are no costs to tolerance traits, tolerance should quickly fix in host plant populations, thereby reducing the selection for resistance alleles (23, 24). When plants are tolerant to pathogen infection, traits that confer resistance are not expected to covary with plant fitness. As a consequence, resistance traits in plants would respond weakly if at all to selection by pathogens. These theoretical models suggest that the extent of variation present for different components of the complex trait of plant resistance, as well as the correlation among components, can alter the evolutionary dynamics of both plant and pathogen. To understand the evolution of plant responses to pathogen infection, the relative importance of resistance and tolerance needs to be investigated empirically. Thus, from an applied perspective, it is important to understand the complex relationships among pathogen growth, plant resistance, and tolerance because not all genes that confer biochemical resistance (in terms of reducing symptoms) will necessarily increase yield (fitness).
In the past decade Arabidopsis thaliana has been developed as a model organism for the study of the mechanism of disease resistance in plants (25). However, fitness consequences of natural variation in disease resistance have not been previously investigated. Here, we investigated heritable variation in resistance to the bacteria Pseudomonas syringae among a worldwide collection of accessions of A. thaliana. In this study, we measured three components of host resistance (pathogen growth, disease symptoms, and host fitness) and determined their covariation to determine the relative importance of resistance and tolerance.
Materials and Methods
Genetic variation in the components of disease resistance was studied in 19 accessions of A. thaliana, described in Table 1. We chose 17 accessions that encompass the geographical range of the species and that include the extremes of A. thaliana genetic diversity based on data from microsatellite and AFLP markers (26, 27). We also included the accessions Col-0 and Ler-0 because they are the accessions commonly used in studies of disease resistance in A. thaliana. Initial seed stocks were obtained from the Arabidopsis Information Management System (www.arabidopis.org). Each accession was grown and selfed for one generation before this study.
Table 1.
List of the accessions used, their stock number at the Arabidopsis Information Management System, and their geographical origin
| Code | Stock | Ecotype | Collection site |
|---|---|---|---|
| 1 | CS6643 | Bur-0 | Ireland |
| 2 | CS6660 | Can-0 | Canary Islands |
| 3 | CS6673 | Col-0 | USA |
| 4 | CS6674 | Ct-1 | Italy |
| 5 | CS6688 | Edi-0 | Scotland |
| 6 | CS6736 | Hi-0 | Netherlands |
| 7 | CS6792 | Kn-0 | Lithuania |
| 8 | CS20 | Ler-0 | Germany |
| 9 | CS1380 | Mt-0 | Libya |
| 10 | CS6805 | No-0 | Germany |
| 11 | CS6824 | Oy-0 | Norway |
| 12 | CS6839 | Po-0 | Germany |
| 13 | CS6850 | Rsch-4 | Russia |
| 14 | CS6857 | Sf-2 | Spain |
| 15 | CS6874 | Tsu-0 | Japan |
| 16 | CS6889 | Wil-2 | Russia |
| 17 | CS6891 | Ws-0 | Russia |
| 18 | CS6897 | Wu-0 | Germany |
| 19 | CS6902 | Zu-0 | Germany |
Plants were grown under uniform conditions and inoculated with the bacteria P. syringae pv. Tomato, strain Pst DC3000 to determine the response to pathogen infection. The bacterial strain Pst DC3000 was chosen because it is a wild-type strain capable of infecting all A. thaliana accessions previously tested (28). Twenty seeds of each accession were planted into four 3-inch pots and vernalized at 4°C for 3 days in the dark. Plants were then grown in a growth chamber under constant temperature (25°C) and humidity (70%) and 8-h photoperiod. After 32 days (before any plant began flowering), two pots of each accession were inoculated by dipping them in a bacterial solution containing 10 mM MgCl2, 0.02% L-77 Silwet, and 107 bacterial cells per ml (29). The other two pots were mock-inoculated by dipping in the above solution without bacterial cells. Plants from the mock-inoculated pots were used as controls.
Five days after inoculation (when symptom expression is at its peak for all accessions) each plant was visually inspected for disease symptoms. Symptoms were scored on a standard scale from 1 (no signs of disease symptoms) to 5 (extensive chlorosis and water-soaked lesions) on inoculated plants. Mock-inoculated plants never presented any symptoms. Bacterial growth was estimated by the number of bacterial cells present per cm2 of leaf tissue. Six disks of 0.25 cm2 area of leaf tissue were collected with a cork borer for each ecotype 5 days after inoculation. Leaf disks were collected randomly in relation to lesions present. The disks were ground in 10 mM MgCl2 solution and plated on NYG agar plates (with 1 mg/ml rifampicin) after appropriate dilutions were made. The number of colony-forming units (CFUs) per plate was counted 24 h later. Three replicate measurements were made for each accession. In each replicate, disks were collected from three different plants of the same accession.
After plants were scored for disease symptoms and leaf disks were collected, day length was gradually increased to 16 h to induce flowering. Plants were kept in the growth chamber until senescence when total fruit production was recorded. Because A. thaliana is an annual plant, the effect of pathogen infection over the lifetime fitness can be estimated by total seed production on senescence. Time to senescence varied among ecotypes, from as early as 1 month to a maximum of 4 months after inoculation. To estimate seed production, three fruits (siliques) from each plant were collected and the number of seeds in each counted. Although there was no statistically significant difference between the number of seeds per fruit produced by different plants of the same accession within treatment, different accessions produced a significantly different number of seeds per fruit. Thus, plant fitness was estimated by multiplying the number of fruits produced per plant by the average number of seeds per fruit for that accession.
To test for natural variation in disease-resistance-related traits among A. thaliana accessions, we used an ANOVA to determine whether accession significantly affects the severity of disease symptoms, the size of bacterial populations in leaves, and seed production after inoculation. Although the use of multiple plants per pot is standard in studies of resistance in A. thaliana, we observed a significant pot effect for all variables. Thus, we present the results from an ANOVA where the effects of pot nested within accession is part of the model, and the main effects are tested with the mean squares for pot nested within accession as the error term. The broad sense heritability (H2) for these three traits was estimated by determining the proportion of the variance explained by the ecotypes from the pot-corrected ANOVA over total variance (30). The effect of P. syringae infection on A. thaliana fitness was determined by comparing the seed production in inoculated and mock-inoculated plants with a hierarchical ANOVA [model: fitness = accession + infection status + (accession × infection) + pot (accession × infection)]. Accession was considered a fixed effect because they were specifically chosen to represent the extremes of the genetic variation present in A. thaliana, they do not represent a random sample of accessions. Infection status was considered fixed and pot was considered a random effect. The correlation between bacterial population size and disease symptoms was determined using Pearson's correlation between the average values of the trait for each accession. To determine the covariance between fitness and symptom severity for each plant we performed a regression analysis. To determine the role of tolerance in the interaction between A. thaliana and P. syringae, we investigated whether the difference in fitness between inoculated and noninoculated plants could be explained by the degree to which the plants become infected (symptoms or bacterial population size). Thus, we performed a regression analysis on the residuals of seed production after the effect of accessions was removed. Using the residuals is necessary to control for differences in seed production among ecotypes when they are not infected.
Results
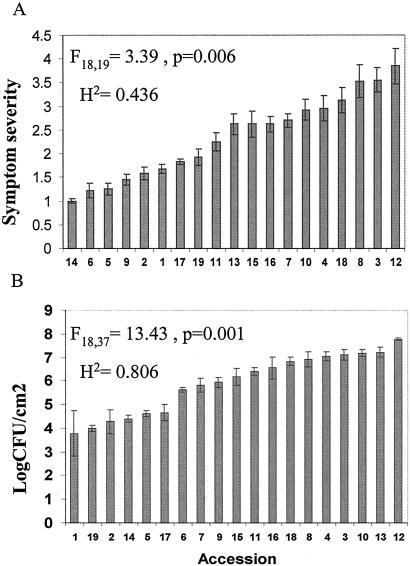
Continuous variation among the accessions of A. thaliana was observed for all three resistance-related traits: the severity of disease symptoms (Fig. 1A), the size of the leaf bacterial population (Fig. 1B), and the fitness of infected plants (Fig. 2). Furthermore, no clear boundaries were detected within the phenotypic distribution that allow classification of accessions as “resistant” or “susceptible.”
Fig 1.
Natural variation in resistance traits to P. syringae among A. thaliana accession. A shows the variation in disease symptoms observed for each accession. Sample sizes for each accession varied between 8 and 10. B shows the variation in the number of bacterial cells detected per cm2 of leaf tissue in each accession. Each bar represents the average of three replicates for each accession and the error bar indicates the standard error of the mean. For details see Materials and Methods.
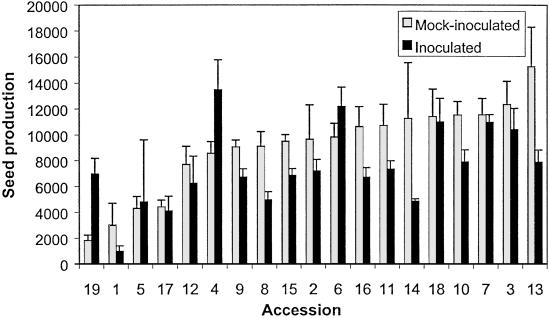
Fig 2.
Effect of P. syringae infection on A. thaliana fitness. Each bar indicates the average number of seeds produced by each A. thaliana accession and the error bar indicates the standard error of the mean. Sample sizes varied between 10 and 7 plants per accession.
The accession with the fewest disease symptoms was Sf-2 (average score 1.0 ± 0.1), whereas the accession exhibiting the greatest disease symptoms was Po-0 (average score 3.9 ± 0.4; Fig. 1). Col-0, which is most widely used for studies of disease resistance in A. thaliana had the second highest average disease score (3.6 ± 0.3). The ANOVA indicated that the 19 accessions differed significantly in the amount of disease symptoms expressed under infection (F18,19 = 3.39; P = 0.006). Variation among accessions explained approximately 44% of the total variance and yields a broad sense heritability estimate of 0.436.
The size of the leaf bacterial population likewise varied among accessions. The accession with the least number of bacteria growing within leaves was Bur-0 (average log number of bacteria 3.78 ± 0.97). Sf-2, which exhibited the fewest disease symptoms, also supported a small bacterial population size (4.40 ± 0.15). The accession with the largest bacterial population, Po-0 (7.77 ± 0.05), also had the highest average score for disease symptoms. Col-0 had the fourth largest average bacterial population size (7.11 ± 0.24). The ANOVA indicated significant differences in the size of the bacterial population present in the leaves of the different accessions (F18,37 = 13.43; P = 3.8 × 10−11). Broad sense heritability was estimated to be 0.806.
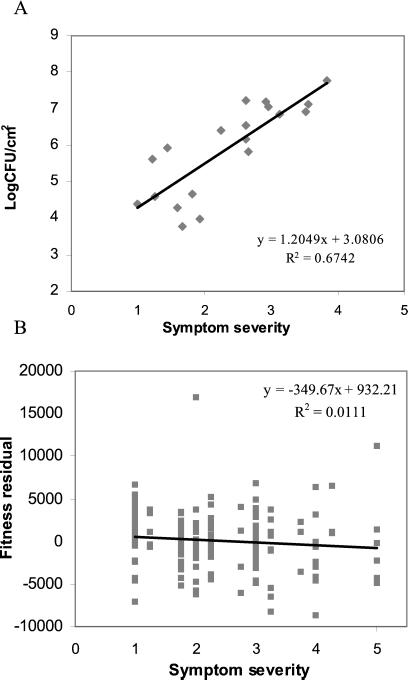
Although bacterial populations were detected in all accessions, some of the accessions showed no disease symptoms at all. It is possible that the expression of symptoms is under threshold control, requiring a minimum number of bacteria within a leaf before symptoms are apparent. Overall, symptom severity and bacterial density are strongly correlated among accessions (Fig. 3A). Thus, both disease symptoms and bacterial growth are equivalent indicators of host resistance.
Fig 3.
Correlation between the three disease resistance-related traits. A shows the correlation between the average number of bacteria growing per cm2 of leaf tissue and symptom severity within each accession. B shows the correlation between symptom severity and fitness under infection for each plant (fitness residuals after difference in seed production due to accession is removed).
To determine the effect of infection on plant fitness, we assessed total seed production for control and infected plants of each accession. The mock-inoculated plants showed significant variation among accessions in total seed production (F18,19 = 2.54; P = 0.025), indicating genetic differences for seed production among accessions. Among mock-inoculated plants, accession Rsch-4 had the highest seed production (on average 15,259 seeds per plant), whereas Zu-1 had the lowest seed production (on average 1,829 seeds per plant). Accessions also differed significantly for seed production when inoculated with P. syringae (F18,19 = 4.03; P = 0.002). Seed production for inoculated plants varied from an average of 977 seeds per plant in Bur-0 to 13,469 seeds per plant in Ct-1 (Fig. 2). Broad sense heritability for seed production under infection was estimated to be 0.292.
The effect of P. syringae infection on A. thaliana fitness was tested with a hierarchical ANOVA to determine the effect of accession, infection status (inoculated vs. control), and the interaction between accession and infection on total seed production. We found that inoculated plants produced on average significantly fewer seeds than control plants (F1,38 = 9.52; P = 0.004). This effect is mediated through both a reduction in the number of seeds per fruit and a reduction in the number of fruits produced per plant (data not shown). We also observed a significant effect of accession on seed production (F18,38 = 9.64; P = 3.4 × 10−9), indicating that accessions differ on their average seed set independent of whether they are infected. Furthermore, the two-way ANOVA revealed a significant interaction between accession and infection (F18,38 = 2.66; P = 0.006), indicating that the effect of infection on seed production varies among accessions (Fig. 2). That is, the reduction in plant fitness caused by infection is not uniform and varies among accessions. This interaction effect can be explained by two different mechanisms. The different fitness response to infection may be simply a consequence of the fact that the accessions harbor bacterial populations of different size because they vary in resistance traits. However, the significant interaction could also have resulted from accessions varying in tolerance—i.e., some plants have higher fitness despite higher degrees of infection because they are more tolerant. If the first hypothesis is correct, we should expect that accessions that have small bacterial populations, like Sf-2, should have a smaller reduction in fitness than accessions that had large bacterial populations and extensive symptoms, like Col-0. However, this is not the case. We found that infection reduced seed production in Col-0 by 16%, whereas in Sf-2 infection reduced seed production by 57%. Among all accessions the correlation between reduction in seed production and bacterial growth (R2 = 0.22; P = 0.37). These results indicate that although infection overall reduces plant fitness, the degree to which a plant becomes infected is not correlated with the amount of fitness lost. Furthermore, these results suggest that the effect of infection on seed production is mediated by different genetic factors than the ones that determine bacterial growth and disease symptoms.
To better understand the relationship between fitness loss and resistance, we calculated the correlation between symptom severity and the fitness residuals after the effect of ecotype on fitness was removed. If all accessions are equally tolerant, and fitness is a direct consequence of how infected a plant becomes, we expect a negative correlation between infection and fitness. In contrast, we found a slightly negative but nonsignificant correlation between the two variables (Fig. 3B). These results indicate that resistance traits in A. thaliana are not good predictors of fitness, and that tolerance traits are playing an important role in mediating plant fitness under infection.
Discussion
Despite the fact that A. thaliana has been the focus of extensive research on the molecular basis of disease resistance, very little is known about the natural quantitative variation in disease resistance and the relationship between biochemical resistance and fitness in this system. Most studies have concentrated on a few accessions and relied on mutagenic-induced variation (refs. 29 and 31, but see ref. 32). Our investigation of natural variation uncovered a remarkable amount of heritable genetic variation among accessions. The variance expressed has a clear quantitative basis, with no clear boundaries between a “resistant” and a “susceptible” group of ecotypes. The extent of variation observed is particularly remarkable because no avirulence genes for A. thaliana have yet been identified in the bacterial strain used (DC3000). Thus, either there are R-genes for DC3000 still unidentified among the ecotypes studied, or the observed variance is mediated by genes other than the classical R-genes (of the gene-for-gene type). These results indicate that understanding the genetic basis of natural variation in disease resistance may require a broader search of traits related to disease resistance than just characterization of R-genes. Although recent efforts have broaden the search for resistance genes that are part of the transduction pathway of R-genes (19, 33, 34), it may be worthwhile to also look for genes directly related to tolerance and to use quantitative genetics approach to investigate genes that underlie natural quantitative variation.
It is commonly assumed that pathogen effect on host fitness is a direct result of pathogen growth in host tissues. Consequently, many empirical studies estimate plant resistance by using a single trait related to disease resistance. The rationale for using a single trait as a surrogate variable to describe disease resistance is based on two assumptions: visual symptoms are a direct consequence of the amount of pathogen present in the host tissue, and pathogen growth/density is directly correlated with the plant biochemical ability to recognize the presence of pathogens and trigger defense. However, there is still limited understanding of the genetic and biochemical pathways connecting pathogen access to host, the expression of symptoms and infection effect on fitness. Moreover, it is not clear whether symptom expression and fitness reduction are part of the same pathway (32, 35). The effect of the pathogen on host evolution depends on whether pathogen infection affects host fitness, but the direction in which hosts will respond to pathogen selection depends on which heritable traits covary with host fitness.
The interaction between A. thaliana and P. syringae is an ideal system to investigate the relationship between pathogen growth, symptoms, and fitness effects because the three variables can be estimated independently. In our study we found extensive heritable quantitative variation among 19 accession of A. thaliana for the three resistance-related traits, indicating that all three traits can respond to pathogen selection. We observed a significant correlation between the size of the bacterial population present in host leaves and the average severity of observed symptoms within each accession. This result supports the idea that disease symptoms are a direct consequence of pathogen growth in host tissues.
Although we observed a significant effect of infection on plant fitness, indicating that pathogens can exert selection on A. thaliana, the effect of infection on fitness varied among accessions. The relative reduction in fitness among the different accessions due to bacterial infection cannot be explained by the intensity of infection (measured as bacterial growth or symptoms), indicating that accessions vary for traits that mediate the effect of disease on fitness—i.e., tolerance traits. Moreover, these results suggest that estimates of pathogen population size and symptoms are not good estimators of pathogen effect on A. thaliana fitness. Because tolerance seems to play an important role in A. thaliana response to P. syringae, and resistance traits do not covary with fitness, we expect that in response to pathogen selection, A. thaliana will respond mainly through tolerance traits.
The role of tolerance (i.e., host's ability to reduce the effect of infection on plant fitness) in plant–pathogen interactions appears to have been mostly overlooked (but see refs. 11 and 12), despite the extensive literature documenting tolerance in plant–herbivore interactions (20). Our study is the first evidence for tolerance in a plant–bacterial interaction and more studies are needed to determine how common and important tolerance is for plant–pathogen interactions. Understanding the role of tolerance is particularly important because theoretical models have shown that tolerance significantly affects plant–pathogen coevolution (23, 24). Furthermore, the existence of significant variation in tolerance can alter the manner in which we search for the genetic basis of traits that can increase yield in crop varieties. Currently most efforts have been directed at understanding the molecular basis of resistance (genes that prevent pathogen establishment or growth; refs. 36–38). Our study suggests that understanding the traits and genes that increase plant tolerance may provide an alternative strategy for reducing crop loss to pathogens.
Acknowledgments
We thank B. Kunkel for technical support, and A. Caicedo, K. Clay, T. Juenger, M. Kramer, B. Kunkel, and J. Wolf for helpful discussions and comments on the manuscript. This work was supported by a National Science Foundation minority postdoctoral fellowship (to P.X.K.).
References
- 1.Pimentel D., Lach, L., Zuniga, R. & Morrison, D. (2000) Bioscience 50, 53-65. [Google Scholar]
- 2.Clay K. & Putten, W. H. V. d. (1999) in Life History Evolution in Plants, eds. Vuorisalo, T. & Mutikainen, P. (Kluwer, Dordrecht, The Netherlands), pp. 275–301.
- 3.Kareiva P. (2000) Proc. Natl. Acad. Sci. USA 96, 8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexander H. M. (1992) in Plant Resistance to Herbivores and Pathogens, eds. Fritz, R. S. & Simms, E. (Univ. of Chicago Press, Chicago), pp. 326–344.
- 5.Leonard K. J. & Levy, Y. (1990) J. Phytopathol. 128, 161-171. [Google Scholar]
- 6.Dingerdissen A. L., Geiger, H. H., Lee, M., Schechert, A. & Welz, H. G. (1996) Mol. Breed. 2, 143-156. [Google Scholar]
- 7.Lewers K. S., Crane, E. H., Bronson, C. R., Schupp, J. M., Keim, P. & Shoemaker, R. C. (1999) Mol. Breed. 5, 33-42. [Google Scholar]
- 8.Parker M. A. (1985) Evolution (Lawrence, Kans.) 39, 713-723. [DOI] [PubMed] [Google Scholar]
- 9.Simms E. L. (1993) Plant Dis. 77, 901-904. [Google Scholar]
- 10.Jarosz A. M. & Burdon, J. J. (1991) Evolution (Lawrence, Kans.) 45, 1618-1627. [DOI] [PubMed] [Google Scholar]
- 11.Simms E. & Triplett, J. (1994) Evolution (Lawrence, Kans.) 48, 1973-1985. [DOI] [PubMed] [Google Scholar]
- 12.Parker M. A. (1986) Oecologia 69, 253-259. [DOI] [PubMed] [Google Scholar]
- 13.Clarke D. D. (1986) Adv. Plant Pathol. 5, 161-197. [Google Scholar]
- 14.Albar L., Lorieux, M., Ahmadi, N., Rimbault, I., Pinel, A., Sy, A. A., Fargette, D. & Ghesquiere, A. (1998) Theor. Appl. Genet. 97, 1145-1154. [Google Scholar]
- 15.Chen S., Porter, P., Orf, J., Reese, C., Stienstra, W., Young, N., Walgenbach, D., Schaus, P., Arlt, T. & Breitenbach, F. (2001) Plant Dis. 85, 760-766. [DOI] [PubMed] [Google Scholar]
- 16.Stout M., Rice, W., Linscombe, S. & Bollich, P. (2001) J. Econ. Entomol. 94, 963-970. [DOI] [PubMed] [Google Scholar]
- 17.Schmid B. (1994) J. Ecol. 82, 165-175. [Google Scholar]
- 18.Burdon J. J., Ericson, L. & Muller, W. J. (1995) J. Ecol. 83, 979-989. [Google Scholar]
- 19.Glazebrook J., Rogers, E. E. & Ausubel, F. M. (1996) Genetics 143, 973-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strauss S. Y. & Agrawal, A. A. (1999) Trends Ecol. Evol. 4, 179-185. [DOI] [PubMed] [Google Scholar]
- 21.Juenger T. & Bergelson, J. (2000) Evolution (Lawrence, Kans.) 54, 764-777. [DOI] [PubMed] [Google Scholar]
- 22.Hamilton W., Axelrod, R. & Tanese, R. (1990) Proc. Natl. Acad. Sci. USA 87, 3566-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roy B. A. & Kirchner, J. W. (2000) Evolution (Lawrence, Kans.) 54, 51-63. [DOI] [PubMed] [Google Scholar]
- 24.Tiffin P. (2000) Am. Nat. 155, 128-138. [DOI] [PubMed] [Google Scholar]
- 25.Kunkel B. (1996) Trends Genet. 12, 63-69. [DOI] [PubMed] [Google Scholar]
- 26.Innan H., Terauchi, R. & Miyashita, N. T. (1997) Genetics 146, 1441-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King G., Nienhuis, D. & Hussey, C. (1993) Theor. Appl. Genet. 86, 1028-1032. [DOI] [PubMed] [Google Scholar]
- 28.Whalen M., Innes, R., Bent, A. & Staskawicz, B. (1991) Plant Cell 3, 49-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kunkel B. N., Bent, A. F., Dahlbeck, D., Innes, R. W. & Staskawicz, B. J. (1993) Plant Cell 5, 865-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitchell-Olds T. & Pedersen, D. (1998) Genetics 149, 739-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alonso-Blanco C. & Koornneef, M. (2000) Trends Plant Sci. 5, 22-29. [DOI] [PubMed] [Google Scholar]
- 32.Glazebrook J. (1999) Curr. Opin. Plant Biol. 2, 280-286. [DOI] [PubMed] [Google Scholar]
- 33.Frye C. & Innes, R. (1998) Plant Cell 10, 947-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morel J. B. & Dangl, J. L. (1999) Genetics 151, 305-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warren R. F., Merritt, P. M., Holub, E. & Innes, R. W. (1999) Genetics 152, 401-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ji C., Smith-Backer, J. & Keen, N. (1998) Curr. Opin. Biotechnol. 9, 202-207. [DOI] [PubMed] [Google Scholar]
- 37.Ellis J., Dodds, P. & Pryor, T. (2000) Curr. Opin. Plant Biol. 3, 278-284. [DOI] [PubMed] [Google Scholar]
- 38.Staskawicz B. J. (2001) Plant Physiol. 125, 73-76. [DOI] [PMC free article] [PubMed] [Google Scholar]