Abstract
The land plants and their immediate green algal ancestors, the charophytes, form the Streptophyta. There is evidence that both the chloroplast DNA (cpDNA) and mitochondrial DNA (mtDNA) underwent substantial changes in their architecture (intron insertions, gene losses, scrambling in gene order, and genome expansion in the case of mtDNA) during the evolution of streptophytes; however, because no charophyte organelle DNAs have been sequenced completely thus far, the suite of events that shaped streptophyte organelle genomes remains largely unknown. Here, we have determined the complete cpDNA (131,183 bp) and mtDNA (56,574 bp) sequences of the charophyte Chaetosphaeridium globosum (Coleochaetales). At the levels of gene content (124 genes), intron composition (18 introns), and gene order, Chaetosphaeridium cpDNA is remarkably similar to land-plant cpDNAs, implying that most of the features characteristic of land-plant lineages were gained during the evolution of charophytes. Although the gene content of Chaetosphaeridium mtDNA (67 genes) closely resembles that of the bryophyte Marchantia polymorpha (69 genes), this charophyte mtDNA differs substantially from its land-plant relatives at the levels of size, intron composition (11 introns), and gene order. Our finding that it shares only one intron with its land-plant counterparts supports the idea that the vast majority of mitochondrial introns in land plants appeared after the emergence of these organisms. Our results also suggest that the events accounting for the spacious intergenic spacers found in land-plant mtDNAs took place late during the evolution of charophytes or coincided with the transition from charophytes to land plants.
It is well recognized that land plants arose from green algae belonging to the Charophyta (1). Land plants and charophytes form the Streptophyta, a lineage sister to the Chlorophyta, which comprises the rest of green algae (2–4). Of the five orders recognized in the Charophyta (5), the Charales have been shown recently to be the closest relatives of land plants (6). A third green-plant lineage, at the base of the split of the Chlorophyta and Streptophyta, is represented possibly by the green alga Mesostigma viride (7–9). This lineage remains controversial, because some phylogenetic analyses placed Mesostigma within the Streptophyta (2, 6, 10). Whatever the exact position of Mesostigma, there is no doubt that this alga belongs to a deeply diverging lineage, because it represents the most basal branch in trees inferred from sequences of land plants and charophytes from all five orders (6).
An understanding of the evolution of chloroplast DNA (cpDNA) and mitochondrial DNA (mtDNA) within the Streptophyta is of great interest, considering that comparative analyses of complete organelle DNA sequences from Mesostigma and land plants highlight considerable differences at the organizational level, with cpDNA showing more conservation than mtDNA (7, 8). Because there exist only fragmentary data on the organization of charophyte organelle DNAs (mainly for the cpDNA of Spirogyra maxima, a member of the Zygnematales; refs. 11–13), it is difficult to predict the timing of the major events that shaped the architectures of land-plant cpDNAs and mtDNAs.
Mesostigma cpDNA is highly similar in size and gene organization to the cpDNAs of the 10 photosynthetic land plants examined thus far (the bryophyte Marchantia polymorpha and nine vascular plants; see www.ncbi.nlm.nih.gov/PMGifs/Genomes/organelles.html), but lacks any introns and contains ≈20 extra genes (7). Chloroplast gene loss is an ongoing process in the Streptophyta, with independent losses occurring in multiple lineages (14, 15). Mesostigma and most land-plant cpDNAs share a quadripartite structure that is characterized by the presence of two copies of a rRNA-containing inverted repeat (IR) separated by large and small single-copy regions. All the genes they have in common, with a few exceptions, reside in corresponding genomic regions, and the great majority are part of conserved clusters.
In contrast, Mesostigma mtDNA greatly differs at the levels of size, gene organization, and intron content from the completely sequenced mtDNAs of the bryophyte (M. polymorpha) and the two angiosperms (Arabidopsis thaliana and Beta vulgaris) that have been investigated thus far (8). During the evolutionary transition from Mesostigma to Marchantia, mtDNA underwent a 4-fold increase in size, was rearranged extensively, and gained many introns while maintaining a similar gene content. After the emergence of bryophytes, mtDNA grew larger via the duplication of noncoding regions and the capture of cpDNA and nuclear DNA sequences, sustained loss of numerous genes, and acquired a highly dynamic genome structure as exemplified by the existence of angiosperm mtDNA as a mixture of recombinational isomers (16, 17). Mesostigma and land-plant mtDNAs share no introns, and only 1 of the 32 introns in Marchantia mtDNA (18) is conserved in A. thaliana (19) and B. vulgaris mtDNAs (20), suggesting that most of the liverwort mitochondrial introns have arisen independently from those present in angiosperms. The distribution patterns of mitochondrial introns among basal land plants are consistent with this hypothesis and also indicate that all five trans-spliced introns conserved among angiosperm mtDNAs arose from cis-spliced intron homologs (21–25). RNA-editing events involving mainly the conversions of cytidine into uridine have been observed in the mitochondria of basal land plants and angiosperms (26–29) as well as in their chloroplasts (26, 30–32) but appear to be absent in both organelles of Marchantia and the few algae examined thus far. These events are more frequent in mitochondria, where they affect essentially every protein-encoding mRNA in angiosperms.
Here, we report the complete cpDNA and mtDNA sequences of the charophyte Chaetosphaeridium globosum, a member of the order (Coleochaetales) that has been identified as the sister group of the Charales and land plants (6). Our comparative analyses of the Chaetosphaeridium organelle genomes with their Mesostigma and land-plant counterparts have allowed us to trace the origins of some of the events that restructured the cpDNA and mtDNA within the Streptophyta.
Materials and Methods
The axenic strain M1311 of C. globosum (Nordstedt) Klebahn was obtained from M. Melkonian (University of Köln, Köln, Germany). Cultures were grown at 18°C under alternating 12-h light/12-h dark periods in medium AF-6 (33). The cpDNA and mtDNA were sequenced in parallel by using a plasmid library prepared from an AT-rich fraction. The methods used were those reported previously (7) except that nucleotide sequences were determined with the PRISM Big Dye terminator cycle-sequencing ready-reaction kit (Applied Biosystems), the PRISM dGTP Big Dye terminator ready-reaction kit (Applied Biosystems), and the DYEnamic ET terminator cycle-sequencing kit (Amersham Pharmacia) on the ABI model 377 DNA sequencer (Applied Biosystems). Sequence analysis was carried out as described (8). The program DERANGE2 (M. Blanchette and D. Sankoff, Université de Montréal, Montréal) and that developed by N. El-Mabrouk and Y. Ajana (Université de Montréal) were used to infer the number of gene permutations by inversions. Maximum-likelihood analysis of combined protein sequences was carried out with PROTML (34) and AAML (35) by using the JTT model and gamma-distributed rates of substitutions across sites. The chloroplast and mitochondrial data sets (11,482 and 4,259 positions, respectively) consisted of the proteins analyzed in refs. 7 and 8, respectively. The homologous sequences of Cyanophora paradoxa were used as outgroup in the chloroplast analysis, whereas those of Porphyra purpurea, Chondrus crispus, and Cyanidioschyzon merolae were used to root the mitochondrial trees.
Results and Discussion
Features of Chaetosphaeridium cpDNA and mtDNA.
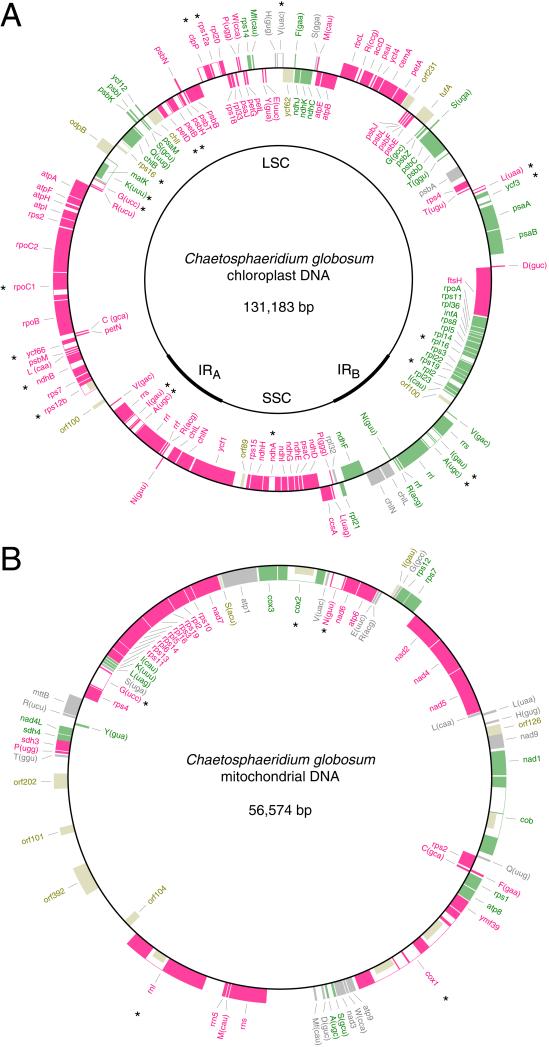
In terms of size, gene content, and intron composition, Chaetosphaeridium cpDNA (131,183 bp) closely resembles Marchantia cpDNA (Table 1 and Fig. 1A). It encodes 124 genes, all of which have been identified in previously investigated green-plant cpDNAs, and features a quadripartite structure in which the IR displays a pair of extra genes (chlL and chlN) as compared with the corresponding genomic region in Marchantia cpDNA. The Chaetosphaeridium chlL/chlN cluster is located at one end of the IR, whereas this cluster is located at the immediate border of the small single-copy region in Marchantia. This observation indicates that expansion/contraction of the IR, a common event in land plants (36), led to the increased size and gene content of the Chaetosphaeridium IR. All the other genes in Chaetosphaeridium cpDNA that are shared with land-plant and Mesostigma cpDNAs are partitioned within the same genomic regions. In contrast, there is no rRNA-encoding IR in the 129.9-kb Spirogyra cpDNA (11). Seventeen group II introns and a unique group I intron are found in Chaetosphaeridium cpDNA; all are similar positionally and structurally to land-plant chloroplast introns. Similar to its land-plant homologs, Chaetosphaeridium cpDNA contains a single cis-spliced intron [in trnK(uuu)] with an ORF (matK) as well as a single trans-spliced intron (rps12.i1 intron). The latter contains an ORF in domain IV, which is missing in land-plant cpDNAs. Before our study, homologs of some land-plant chloroplast introns had been identified in other charophytes (12, 13, 37).
Table 1.
General features of cpDNA
| Feature | Mesostigma | Chaetosphaeridium | Marchantia | Arabidopsis |
|---|---|---|---|---|
| Size, bp | 118,360 | 131,183 | 121,024 | 154,478 |
| A + T content, % | 69.9 | 70.4 | 71.2 | 63.7 |
| Gene content | 136 | 125 | 120 | 110 |
| Introns | ||||
| Group I | 0 | 1 | 1 | 1 |
| Group II | ||||
| cis-spliced | 0 | 16 | 18 | 19 |
| trans-spliced | 0 | 1 | 1 | 1 |
Unique ORFs, intron ORFs, and pseudogenes were not taken into account.
Fig 1.
Gene maps of Chaetosphaeridium cpDNA (A) and mtDNA (B). Genes outside each map are transcribed clockwise. Genes absent from Marchantia cpDNA and mtDNA are represented in beige. Gene clusters shared with Marchantia are shown as series of green and red boxes. Genes present in Marchantia but located outside conserved clusters are shown in gray. tRNA genes are indicated by the one-letter amino acid code followed by the anticodon in parentheses. Intron-containing genes are denoted by asterisks, with the introns represented as open boxes. The intron sequences bordering the rps12 exons (rps12a and rps12b) are spliced in trans at the RNA level.
Chaetosphaeridium mtDNA (56,574 bp) is more alike Mesostigma mtDNA than Marchantia mtDNA at the levels of size and intron composition (Table 2). Its 67 genes, all previously identified in green-plant mtDNAs, are tightly packed in a 48.3-kb segment, the gene density of which is comparable to that found in Mesostigma mtDNA (Fig. 1B). The remaining 8.3-kb segment mainly accounts for the increased size of Chaetosphaeridium mtDNA relative to its Mesostigma homolog. Two of the four ORFs found in this segment, those lying at the borders and on opposite strands (orf101 and orf202), show sequence homology with phage integrase/recombinase genes and with orf304 in the mtDNA of the chlorophyte Prototheca wickerhamii. Given this observation and the fact that none of the potential coding sequences of the 8.3-kb segment are commonly found in mtDNA, it is possible that this segment took residence in mtDNA via horizontal transfer of phage or bacterial DNA. Nine group I introns and two group II introns were identified in six mitochondrial genes of Chaetosphaeridium. Only one (cox1.i2) of these introns is homologous positionally and structurally to an intron in land-plant mtDNAs (in Marchantia mtDNA), and only one (cox1.i4) has a known homolog in Mesostigma mtDNA (Table 3).
Table 2.
General features of mtDNA
| Feature | Mesostigma | Chaetosphaeridium | Marchantia | Arabidopsis |
|---|---|---|---|---|
| Size, bp | 42,424 | 56,574 | 186,609 | 366,924 |
| A + T content, % | 67.8 | 65.6 | 57.6 | 55.2 |
| Coding sequences, % | 86.6 | 76.3 | 65.0 | 45.5 |
| Gene content | 65 | 67 | 69 | 50 |
| Introns | ||||
| Group I | 4 | 9 | 7 | 0 |
| Group II | ||||
| cis-spliced | 1 | 2 | 25 | 18 |
| trans-spliced | 2 | 0 | 0 | 5 |
For Mesostigma mtDNA, the data were taken from ref. 8, whereas those for the Marchantia and Arabidopsis mtDNAs were taken from ref. 47.
Conserved genes, unique ORFs, introns, and intron ORFs were considered as coding sequences.
Unique ORFs, intron ORFs, and pseudogenes were not taken into account.
Table 3.
Chaetosphaeridium mitochondrial introns
| Intron | Group | ORF size, codons | ORF location | Source of homologous introns |
|---|---|---|---|---|
| cob.i1 | ID | — | — | Nephroselmis olivacea mt (i1) |
| Chlamydomonas smithii mt (i1) | ||||
| cob.i2 | ID | 207 | L2 | Saccharomyces cerevisiae mt (i2) |
| coxl.i1 | IB2 | — | — | Allomyces macrogynus mt (i2) |
| Emericella nidulans mt (i1) | ||||
| Podospora anserina mt (i3) | ||||
| coxl.i2 | IB4 | 303 | L8 | Agrocybe aegerita mt (i1) |
| Allomyces macrogynus mt (i5) | ||||
| Marchantia polymorpha mt (i4) | ||||
| Prototheca wickerhamii mt (i1) | ||||
| Saccharomyces douglasii mt (i1) | ||||
| Schizosaccharomyces pombe mt (i1) | ||||
| coxl.i3 | IB1 | — | — | Podospora anserina mt (i11) |
| coxl.i4 | IB2 | — | — | Allomyces macrogynus mt (i8) |
| Chlorogonium elongatum mt (i1) | ||||
| Mesostigma viride mt (i1) | ||||
| Podospora anserina mt (i12) | ||||
| coxl.i5 | IB2 | 267 | L6 | Podospora anserina mt (i14) |
| cox2.i1 | ID | 269 | L2 | Dictyostelium discoideum mt (i4) |
| rnl.i1 | IB4 | 170 | L6 | Chlorosarcina brevispinosa cp |
| trnG(ucc).i1 | IIA | — | — | — |
| trnN(guu).i1 | — | — | — |
L followed by a number refers to the loop extending the base-paired region identified by the number. Each of the ORFs identified potentially codes for a protein with the LAGLIDADG motif.
Introns inserted at identical gene locations (mt, mitochondria; cp, chloroplast).
Insertion site corresponding to position 1917 in Escherichia coli 23S rRNA.
The absence of domain VI, which carries the highly conserved bulged adenosine that acts as the nucleophile in the first step of splicing by transesterification, prevented the determination of the sequence signatures that distinguish IIA and IIB introns. It is possible that splicing of this intron proceeds through a pathway in which water is the nucleophile (50).
Patterns of Gene Losses in Streptophyte cpDNAs and mtDNAs.
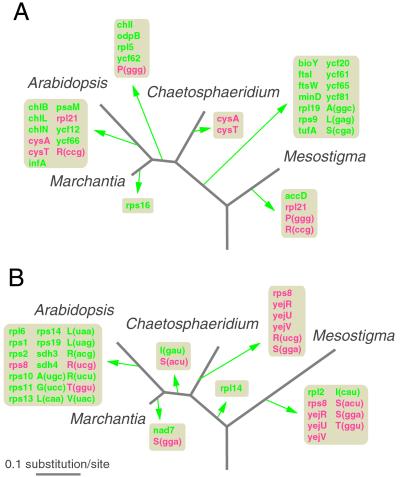
We compared the gene contents of Mesostigma (7, 8), Chaetosphaeridium, Marchantia (18, 38), and Arabidopsis (19, 39) organelle DNAs and inferred the events of gene loss from the cpDNA and mtDNA by using the single-tree topology that was retrieved after separate maximum-likelihood analyses of the combined protein sequences predicted from 53 chloroplast genes and 19 mitochondrial genes (Fig. 2). Sequences from chlorophyte taxa were not included in the data sets, because the current controversy regarding the position of Mesostigma relative to chlorophytes could not be resolved by the addition of the Chaetosphaeridium taxon. When chlorophyte sequences were included in the data sets, trees based on chloroplast proteins identified Mesostigma as the earliest green-plant divergence, but mitochondrial trees gave low support for this position (data not shown). All phylogenetic analyses unambiguously placed Chaetosphaeridium at the base of land-plant lineages.
Fig 2.
Phylogenetic distribution of gene loss from cpDNA (A) and mtDNA (B) in the Streptophyta. The genes that were lost independently in different lineages are indicated in red. The predicted genes in the organelle DNAs of the common ancestor of Mesostigma, Chaetosphaeridium, and land plants are listed in Tables 4 and 5. In the lineage leading to Arabidopsis, the tRNA genes that were lost from mtDNA after the insertion of homologous cpDNA sequences (17) are not indicated. The chloroplast and mitochondrial trees were drawn on the same scale. For both phylogenetic analyses, the Arabidopsis protein sequences were predicted from the corresponding gene sequences without taking into account the edited sites in mRNA.
Of the 140 genes predicted to have been present in the cpDNA of the last common ancestor of Mesostigma and Chaetosphaeridium (Table 4, which is published as supporting information on the PNAS web site, www.pnas.org), 19 were lost during the transition from this ancestor to the common ancestor of all land plants, with 14 of these loss events recorded during the interval separating the Mesostigma and Chaetosphaeridium lineages (Fig. 2A). We included tufA among the genes that were lost, although a sequence homologous to this gene was detected in Chaetosphaeridium cpDNA. Similar to the tufA sequence in the cpDNA of another member of the Coleochaetales (Coleochaete orbicularis; ref. 40), that of Chaetosphaeridium is highly divergent from those of other green plants and most probably codes for a nonfunctional protein. Because tufA is present in the nuclear genome of Arabidopsis (41) and also because tufA-like sequences have been detected by hybridization in the nuclear DNA of certain charophytes (including Coleochaete; ref. 40), the function of this gene in the Coleochaetales most probably has been replaced by a copy of the same gene that was transferred to the nucleus early during charophyte evolution (40).
Mitochondrial genes were lost at a significantly lower frequency than chloroplast genes during the transition from the last common ancestor of Mesostigma and Chaetosphaeridium to the common ancestor of all land plants (Fig. 2B). Only three of the 74 mitochondrial genes predicted in the former ancestor (Table 5, which is published as supporting information on the PNAS web site) were lost during this evolutionary period. Conversely, a higher proportion of mitochondrial genes suffered loss in the lineages leading to Mesostigma and Chaetosphaeridium, with five genes lost independently in these two lineages.
Changes in Chloroplast and Mitochondrial Gene Orders in Streptophyta.
The similarity in gene order between Chaetosphaeridium and Marchantia cpDNAs is remarkable. These cpDNAs share 12 blocks of colinear sequences (Fig. 1A), and up to 40 genes are present in individual blocks, the average number being 10. Only six genes, four of which are tRNA genes, are not comprised within common blocks. The differences in gene order in the small single-copy region can be explained by a single inversion, whereas those in the large single-copy region are attributable to 11 inversions that share 7 endpoints. As in rearranged land-plant cpDNAs (42, 43), tRNA genes often occur at the rearrangement breakpoints (at 12 of the 17 different endpoints identified), suggesting that chloroplast gene order in both charophyte and land-plant cpDNAs is scrambled by recombination across short indirect repeats found within or near tRNA genes. In contrast, Chaetosphaeridium cpDNA differs from its Mesostigma homolog by many rearrangements. These cpDNAs share 24 blocks of colinear sequences, each containing four genes in average, and 44 inversions (7 in the small single-copy region and 37 in the large single-copy region) account for the observed structural differences. Interestingly, a smaller number (37) of inversions are required for converting the gene order of Marchantia cpDNA into that of Mesostigma cpDNA; this observation is consistent with our finding that Chaetosphaeridium cpDNA has not retained some of the ancestral gene clusters shared by Mesostigma and land-plant cpDNAs (Table 6, which is published as supporting information on the PNAS web site). We find an identical gene order in the Chaetosphaeridium, Marchantia, and Mesostigma IRs when we disregard the differences in gene content arising from the expansion/contraction of the IR. As judged from the few genes that have been mapped on S. maxima cpDNA (11), this genome is rearranged more extensively than its Chaetosphaeridium homolog relative to Mesostigma and land-plant cpDNAs, supporting the hypothesis that the IR stabilizes the structure of cpDNA by decreasing intramolecular recombination (44).
Gene order has been preserved also in Chaetosphaeridium and Marchantia mtDNAs but to a lesser extent than in the cpDNAs of these green plants. Fifteen blocks of colinear mitochondrial sequences containing three genes on average were identified (Fig. 1B). This level of conservation was unexpected given that all previously analyzed green-plant mtDNAs bear little resemblance with one another at the gene-order level. Only two pairs of genes (rpl6-rps13 and rps12-rps7) are shared between Chaetosphaeridium and Mesostigma mtDNAs. A total of 27 inversions (using 34 different endpoints) would be required to convert the gene order of Chaetosphaeridium mtDNA into that of Marchantia mtDNA, whereas 59 and 61 inversions account for the gene rearrangements displayed by the Mesostigma/Marchantia and Chaetosphaeridium/Mesostigma mtDNA pairs, respectively. As aforementioned for cpDNA, a large fraction of the inversion endpoints (27/34) inferred in the Chaetosphaeridium/Marchantia mtDNA comparison is associated with tRNA genes.
Patterns of Intron Gains and Losses in Streptophyte cpDNAs and mtDNAs.
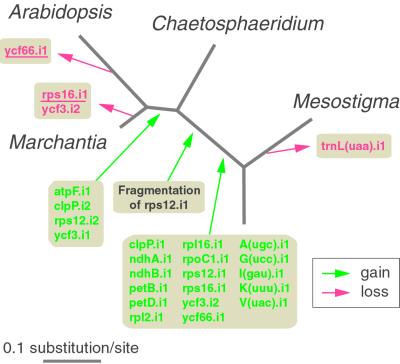
The single group I intron in Chaetosphaeridium and land-plant cpDNAs, which resides in trnL(uaa), is believed to be the most ancient intron found yet. Phylogenetic analyses (37, 45) indicate that it was present in the cyanobacterial ancestor of all chloroplasts and that it originated early during the evolution of cyanobacteria; its absence from some algal cpDNAs (e.g., Mesostigma cpDNA) is attributed to independent losses from trnL(uaa). With regards to the origin of the 21 different group II introns identified in land-plant cpDNAs, the pattern of intron distribution among green plants (Table 7, which is published as supporting information on the PNAS web site) predicts that 17 of these introns were acquired during the interval separating the Mesostigma and Chaetosphaeridium lineages (Fig. 3). Considering that trans-spliced introns have been shown to evolve from cis-spliced orthologs (23, 24), the gain of a cis-spliced version of the trans-spliced rps12.i1 intron must have preceded fragmentation of the intron and relocalization of the resulting intron–exon pieces. Available cpDNA sequences from zygnematalean charophytes (9, 12, 13) including Spirogyra indicate that the latter events as well as the insertions of the trnI(gau) and trnA(ugc) introns occurred before the emergence of the Zygnematales. The four group II introns missing from Chaetosphaeridium cpDNA might have been acquired by streptophytes after the emergence of the Chaetosphaeridium lineage, or alternatively, they might have been gained earlier and lost subsequently. It is possible that proliferation of a few founding group II introns gave rise to the large intron population found in Chaetosphaeridium and land-plant cpDNAs. Analysis of cpDNAs from other charophyte lineages would be helpful in testing this hypothesis.
Fig 3.
Phylogenetic distribution of chloroplast intron gain and loss in the Streptophyta. The chloroplast tree presented in Fig. 2 is shown. The introns that are indicated in red and are underlined were lost from cpDNA together with the genes they interrupt.
The pattern of intron distribution in land-plant mtDNAs together with our finding that Chaetosphaeridium mtDNA shares only one intron (cox1.i2) with its land-plant counterparts are consistent with, but do not prove, the idea that the great majority of land-plant mitochondrial introns originated after the emergence of land plants. Analysis of mtDNAs from other charophyte lineages, especially the Charales, would be required to identify the complete subset of Marchantia and/or angiosperm mitochondrial introns that took their origin during charophyte evolution. The cox1.i4 intron in Chaetosphaeridium mtDNA seems to have been inherited by vertical descent from a common ancestor of Mesostigma and charophytes. The remaining 10 Chaetosphaeridium introns likely were gained through horizontal transfers, because homologs of all these introns, except the two group II introns interrupting tRNA genes, have been identified in the mtDNAs of distantly related eukaryotes, mostly fungi and chlorophyte green algae (Table 3).
Prediction of Protein Sequence Alignments About RNA Editing in Chaetosphaeridium Organelles.
The predicted proteins encoded by Chaetosphaeridium organelle genes were compared with their homologs in plants, algae, and bacteria. No unusual amino acids were identified at sites that are known to be edited in tobacco, rice, and maize chloroplast ORFs (46) as well as in Arabidopsis mitochondrial ORFs (29), and outside these sites only exceptional ones revealed potential mismatch corrections that could be corrected by C-to-U or U-to-C editing. Therefore, our analyses provide no compelling evidence for the occurrence of RNA editing in Chaetosphaeridium organelle transcripts. Sequence analysis of the RNA population in Chaetosphaeridium chloroplasts and mitochondria would be necessary to reject with confidence the possibility that scarce sites are edited.
Concluding Remarks.
By disclosing details on the cpDNA and mtDNA architectures of a charophyte, our study provides a better understanding of how organelle genomes have evolved in the Streptophyta. We have shown that the chloroplast genome acquired the features characteristic of land-plant lineages at an earlier stage than the mitochondrial genome. At various levels, Chaetosphaeridium cpDNA is strikingly similar to land-plant cpDNAs; however, Chaetosphaeridium mtDNA is more compact than its land-plant relatives and features a very different intron composition. The events that gave rise to the spacious intergenic spacers found in land-plant mtDNAs took place either late during the evolution of charophytes or coincided with the transition from charophytes to land plants.
Supplementary Material
Acknowledgments
We thank Michael Melkonian for kindly donating an axenic strain of Chaetosphaeridium. We also thank Nadia El-Mabrouk and Yasmine Ajana for help with the analysis of genome rearrangements. M.T. and C.L. are associates in the Program in Evolutionary Biology of the Canadian Institute for Advanced Research. This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada.
Abbreviations
cpDNA, chloroplast DNA
mtDNA, mitochondrial DNA
IR, inverted repeat
References
- 1.Graham L. E., Cook, M. E. & Busse, J. S. (2000) Proc. Natl. Acad. Sci. USA 97, 4535-4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marin B. & Melkonian, M. (1999) Protist 150, 399-417. [DOI] [PubMed] [Google Scholar]
- 3.Chapman R. L., Buchheim, M. A., Delwiche, C. F., Friedl, T., Huss, V. A., Karol, K. G., Lewis, L. A., Manhart, J., McCourt, R. M., Olsen, J. L. & Waters, D. A. (1998) in Molecular Systematics of Plant II DNA Sequencing, eds. Soltis, D. E., Soltis, P. S. & Doyle, J. J. (Kluwer, Norwell, MA), pp. 508–540.
- 4.Friedl T. (1997) Plant Syst. Evol. Suppl. 11, 87-101. [Google Scholar]
- 5.Mattox K. R. & Stewart, K. D. (1984) in Systematics of the Green Algae, eds. Irvine, D. E. G. & John, D. M. (Academic, London), pp. 29–72.
- 6.Karol K. G., McCourt, R. M., Cimino, M. T. & Delwiche, C. F. (2001) Science 294, 2351-2353. [DOI] [PubMed] [Google Scholar]
- 7.Lemieux C., Otis, C. & Turmel, M. (2000) Nature (London) 403, 649-652. [DOI] [PubMed] [Google Scholar]
- 8.Turmel M., Otis, C. & Lemieux, C. (2002) Mol. Biol. Evol. 19, 24-38. [DOI] [PubMed] [Google Scholar]
- 9.Turmel M., Ehara, M., Otis, C. & Lemieux, C. (2002) J. Phycol. 38, 364-375. [Google Scholar]
- 10.Bhattacharya D., Weber, K., An, S. S. & Berning-Koch, W. (1998) J. Mol. Evol. 47, 544-550. [DOI] [PubMed] [Google Scholar]
- 11.Manhart J. R., Hoshaw, R. W. & Palmer, J. D. (1990) J. Phycol. 26, 490-494. [Google Scholar]
- 12.Lew K. A. & Manhart, J. R. (1993) J. Phycol. 29, 500-505. [Google Scholar]
- 13.Manhart J. R. & Palmer, J. D. (1990) Nature (London) 345, 268-270. [DOI] [PubMed] [Google Scholar]
- 14.Martin W., Stoebe, B., Goremykin, V., Hansmann, S., Hasegawa, M. & Kowallik, K., V. (1998) Nature (London) 393, 162-165. [DOI] [PubMed] [Google Scholar]
- 15.Turmel M., Otis, C. & Lemieux, C. (1999) Proc. Natl. Acad. Sci. USA 96, 10248-10253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmer J. D., Adams, K. L., Cho, Y., Parkinson, C. L., Qiu, Y. L. & Song, K. (2000) Proc. Natl. Acad. Sci. USA 97, 6960-6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marienfeld J., Unseld, M. & Brennicke, A. (1999) Trends Plant Sci. 4, 495-502. [DOI] [PubMed] [Google Scholar]
- 18.Oda K., Yamato, K., Ohta, E., Nakamura, Y., Takemura, M., Nozato, N., Akashi, K., Kanegae, T., Ogura, Y., Kohchi, T. & Ohyama, K. (1992) J. Mol. Biol. 223, 1-7. [DOI] [PubMed] [Google Scholar]
- 19.Unseld M., Marienfeld, J. R., Brandt, P. & Brennicke, A. (1997) Nat. Genet. 15, 57-61. [DOI] [PubMed] [Google Scholar]
- 20.Kubo T., Nishizawa, S., Sugawara, A., Itchoda, N., Estiati, A. & Mikami, T. (2000) Nucleic Acids Res. 28, 2571-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiu Y. L., Cho, Y., Cox, J. C. & Palmer, J. D. (1998) Nature (London) 394, 671-674. [DOI] [PubMed] [Google Scholar]
- 22.Pruchner D., Nassal, B., Schindler, M. & Knoop, V. (2001) Mol. Genet. Genomics 266, 608-613. [DOI] [PubMed] [Google Scholar]
- 23.Malek O. & Knoop, V. (1998) RNA 4, 1599-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malek O., Brennicke, A. & Knoop, V. (1997) Proc. Natl. Acad. Sci. USA 94, 553-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beckert S., Steinhauser, S., Muhle, H. & Knoop, V. (1999) Plant Syst. Evol. 218, 179-192. [Google Scholar]
- 26.Maier R. M., Zeltz, P., Kossel, H., Bonnard, G., Gualberto, J. M. & Grienenberger, J. M. (1996) Plant Mol. Biol. 32, 343-365. [DOI] [PubMed] [Google Scholar]
- 27.Malek O., Lattig, K., Hiesel, R., Brennicke, A. & Knoop, V. (1996) EMBO J. 15, 1403-1411. [PMC free article] [PubMed] [Google Scholar]
- 28.Steinhauser S., Beckert, S., Capesius, I., Malek, O. & Knoop, V. (1999) J. Mol. Evol. 48, 303-312. [DOI] [PubMed] [Google Scholar]
- 29.Giegé P. & Brennicke, A. (1999) Proc. Natl. Acad. Sci. USA 96, 15324-15329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freyer R., Kiefer-Meyer, M. C. & Kossel, H. (1997) Proc. Natl. Acad. Sci. USA 94, 6285-6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirose T., Kusumegi, T., Tsudzuki, T. & Sugiura, M. (1999) Mol. Gen. Genet. 262, 462-467. [DOI] [PubMed] [Google Scholar]
- 32.Yoshinaga K., Iinuma, H., Masuzawa, T. & Uedal, K. (1996) Nucleic Acids Res. 24, 1008-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kato S. (1982) Jpn. J. Phycol. 30, 63-67. [Google Scholar]
- 34.Adachi J. & Hasegawa, M. (1996) Comput. Sci. Monogr. 28, 1-150. [Google Scholar]
- 35.Yang Z. (1997) Comput. Appl. Biosci. 13, 555-556. [DOI] [PubMed] [Google Scholar]
- 36.Goulding S. E., Olmstead, R. G., Morden, C. W. & Wolfe, K. H. (1996) Mol. Gen. Genet. 252, 195-206. [DOI] [PubMed] [Google Scholar]
- 37.Besendahl A., Qiu, Y. L., Lee, J., Palmer, J. D. & Bhattacharya, D. (2000) Curr. Genet. 37, 12-23. [DOI] [PubMed] [Google Scholar]
- 38.Ohyama K., Fukuzawa, H., Kohchi, T., Shirai, H., Sano, T., Sano, S., Umesono, K., Shiki, Y., Takeuchi, M., Chang, Z., et al. (1986) Nature (London) 322, 572-574. [Google Scholar]
- 39.Sato S., Nakamura, Y., Kaneko, T., Asamizu, E. & Tabata, S. (1999) DNA Res. 6, 283-290. [DOI] [PubMed] [Google Scholar]
- 40.Baldauf S. L., Manhart, J. R. & Palmer, J. D. (1990) Proc. Natl. Acad. Sci. USA 87, 5317-5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baldauf S. L. & Palmer, J. D. (1990) Nature (London) 344, 262-265. [DOI] [PubMed] [Google Scholar]
- 42.Cosner M. E., Jansen, R. K., Palmer, J. D. & Downie, S. R. (1997) Curr. Genet. 31, 419-429. [DOI] [PubMed] [Google Scholar]
- 43.Palmer J. D. (1985) in Monographs in Evolutionary Biology: Molecular Evolutionary Genetics, ed. MacIntyre, R. J. (Plenum, New York), pp. 131–240.
- 44.Palmer J. D. (1991) in The Molecular Biology of Plastids, eds. Bogorad, L. & Vasil, K. (Academic, San Diego), pp. 5–53.
- 45.Paquin B., Kathe, S. D., Nierzwicki-Bauer, S. A. & Shub, D. A. (1997) J. Bacteriol. 179, 6798-6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsudzuki T., Wakasugi, T. & Sugiura, M. (2001) J. Mol. Evol. 53, 327-332. [DOI] [PubMed] [Google Scholar]
- 47.Gray M. W., Lang, B. F., Cedergren, R., Golding, G. B., Lemieux, C., Sankoff, D., Turmel, M., Delage, E., Littlejohn, T. G., Plante, I., et al. (1998) Nucleic Acids Res. 26, 865-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Michel F. & Westhof, E. (1990) J. Mol. Biol. 216, 585-610. [DOI] [PubMed] [Google Scholar]
- 49.Michel F., Umesono, K. & Ozeki, H. (1989) Gene 82, 5-30. [DOI] [PubMed] [Google Scholar]
- 50.Podar M., Chu, V. T., Pyle, A. M. & Perlman, P. S. (1998) Nature (London) 391, 915-918. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.