Abstract
There is a great difference in susceptibility to v-abl transgene-induced plasmacytoma between the BALB/cAn and the relatively resistant C57BL/6J mouse strains. We have used the Mapmaker/SURVIVOR algorithm to analyze genome-wide scans on over 800 transgenic F2 hybrid mice, and have mapped at least six loci on chromosomes 2, 4, 11, 17, and 18 that modify tumor-related morbidity. As in human multiple myeloma, males were found to be more prone to plasmacytomagenesis. Different loci influence tumor susceptibility in male and female mice. Survival in females may be largely controlled by a pair of interacting loci on chromosomes 2 and 17.
Genetic control of cancer susceptibility is complex; some common malignancies are significantly heritable (1), and yet the penetrance of known inherited tumorigenic mutations is limited (2). The lower the penetrance of novel tumor modifier genes, the more necessary are models using inbred rodents for gene mapping and identification.
Mouse plasmacytoma and human multiple myeloma are tumors of plasma cells, the end stage of Ig-secreting B-lymphoid cell differentiation. The most extensively investigated mouse model is induction by i.p. pristane, which has been used to implicate the Ink4a-Arf (Cdkn2a) gene (3, 4) and one other tumor modifier (5) on chromosome 4 by linkage mapping studies. Notwithstanding these successes, the pristane model has not been used to address one of the characteristics of the human disease; males are more susceptible than females (6).
An alternative plasmacytoma model is provided by EμSV-v-abl transgenic mice, in which expression of the v-abl protein tyrosine kinase, the oncogene product of the Abelson murine leukemia virus, is controlled by the Ig heavy chain enhancer (Eμ) and a simian virus 40 (SV40) T antigen promoter (7). The transgene is not directly tumorigenic, but in its original (C57BL/6 × SJL/J)F2 genetic context it strongly predisposed mice to develop plasmacytoma, presumably as a result of spontaneous somatic mutations that cooperated with the v-abl oncogene to transform plasma cells to malignancy. Backcross experiments have shown that the v-abl transgene insertion under consideration here continues to produce exclusively plasmacytomas in BALB/c and C57BL/6 mice, and furthermore, has revealed BALB/c mice to be more susceptible than C57BL/6 mice (8).
Here we report analysis of the tumor morbidity free survival of inbred strains and their F1 hybrids, and a linkage analysis of the genetic control of v-abl transgene-dependent plasmacytoma. Whole genome allelic analyses were performed on more than 800 (BALB/c × C57BL/6)F2 animals carrying the v-abl transgene. Linkage was sought to the phenotype of survival time until terminal illness because of tumor. For this purpose, we developed an analytic tool, Mapmaker/SURVIVOR, which is a linkage algorithm specifically designed for analyzing survival data. It is similar to Mapmaker/QTL (9) in providing interval mapping and logarithm of odds (LOD) scores based on an expectation-maximization algorithm allowing most complete use of available data. Mapmaker/SURVIVOR provides increased power obtained from parametric testing compared with nonparametric analyses, and appropriate handling of censored data. Its LOD scores are generated under the Cox proportional hazards model.
Different linkage algorithms were used to detect different features of the data. Single-locus effects were determined by using Mapmaker/SURVIVOR. Locus–locus interactions were investigated by using binary regression trees. Suggestive linkage to the chromosome 4 loci important in the pristane system (10, 11) was obtained. Of greater importance in this system are five or more additional loci, some of which are sex specific. There appears to be at least one substantial epistatic interaction.
Materials and Methods
Mouse Breeding.
EμSV-v-abl 40 (SJL/JWehi × C57BL/6J)F2 animals (7) were backcrossed against both the BALB/cAnBradleyWehi and the C57BL/6J strains while maintaining transgene heterozygosity. When the intercrosses described here were commenced, the C57BL/6 line (B6.abl40) had reached the 12th generation of backcrossing, and the BALB/c line (C.abl40) had reached the 28th generation.
Heterozygous transgene positive B6.abl40 mice were mated with BALB/c mice to produce an F1 generation consisting of transgene heterozygous and nontransgenic mice in approximately equal proportions. Transgene-positive F1 mice were mated with transgene-negative F1 mice to produce an F2 generation, likewise consisting of both transgene-heterozygous and transgene-negative mice. Mice that survived longer than 1 year and that initially had been typed as transgene positive were retyped before linkage analyses were performed.
Animal Husbandry.
Mice were fed γ-irradiated mouse breeder ration (Barastoc Stock Feeds) and provided with acidified drinking water ad libitum. The mouse facilities had a 14 h light/10 h dark cycle and were kept at approximately 22°C. The C.abl40 and B6.abl40 lines and the F1 mice were maintained in a specific pathogen free (SPF) facility. An open access mouse facility was used for most of the F2 mice.
All mice were inspected daily and palpated for abdominal tumor masses weekly. Mice were killed for autopsy on evidence of clinical illness. Autopsies performed on B6.abl 40 and F2 mice surviving to 1 year revealed macroscopic tumor in ≈2% of them. Tissue samples from all sick mice were fixed for histological processing and staining of sections with hematoxylin and eosin by conventional methods. Serum samples taken immediately before killing the sick mice were electrophoresed for the detection of paraproteins as described (7).
Transgene Genotyping.
DNA was purified by standard methods from tail tips or liver samples and stored in TE buffer (10 mM Tris/1 mM EDTA, pH 8.0). Genotyping was performed by using primers within the SV40 late region polyadenylation signal sequence: ggAACTgATgAATgggAgCAgTgg and gCAgACACTCTATgCCTgTgTgg. PCRs contained Taq polymerase, rabbit antiserum to Taq polymerase, 50 mM KCl, 10 mM Tris⋅HCl (pH 8.3), 2.0 mM MgCl2, 0.125 mM deoxynucleoside triphosphates, and 0.14 μM oligonucleotide primers.
Comparison of Transgene Expression.
Ten-microgram samples of total RNA isolated from femoral bone marrow cells of 6-week-old mice by using RNAgents (Promega) were fractionated on 1% formaldehyde-agarose gels and transferred onto Hybond C membranes (Amersham Pharmacia). Filters were baked, prehybridized in 50% HCHO/5× SSC/0.02% wt/vol bovine albumin/500 μg/ml denatured herring sperm DNA, and then hybridized for 18 h at 42°C with probes at 2 × 106 cpm/ml. Filters were washed in 0.2× SSC/0.1% SDS at 65°C and autoradiographed at −70°C. The probes used were a 238-bp Sau3A/BamHI fragment spanning the SV40 early region polyadenylation sequence (7) and a 1.1-kb PstI rat GAPDH cDNA (12), both radiolabeled with [α-32P]dATP to 5 × 108 to 109 cpm/μg. Autoradiography exposures for these probes were 120 and 24 h, respectively.
Genotyping.
The entire transgene positive F2 generation was subjected to genotyping of the autosomes and X chromosome using 191 informative microsatellite markers from the MIT map (13–15) polymorphic between BALB/c and C57BL/6. Markers were placed at an average separation of 10 centimorgans. The microsatellites were amplified by touchdown PCR using forward primers labeled with FAM, HEX, or NED fluorescent dye. PCRs contained Taq polymerase, rabbit antiserum to Taq polymerase, 50 mM KCl, 10 mM Tris⋅HCl (pH 8.3), 2.5 mM MgCl2, 0.125 mM deoxynucleoside triphosphates, and 0.08 μM of the forward and reverse oligonucleotide primers. Reaction products with compatible allele sizes were pooled. Four microliters of pooled product was added to 0.23 μl Genescan 500-ROX DNA size standard (Applied Biosystems), 0.31 μl loading buffer (Applied Biosystems), and 1.6 μl deionized spectral grade formamide (Eastman Kodak). This mixture was heated to 95°C for 5 min before denaturing polyacrylamide gel electrophoretic separation using ABI 377 sequenators. Fluorescence data were collected by using ABI PRISM 377 Collection software. Standard curve fitting and feature analysis were performed by using genescan (Applied Biosystems) software. Alleles were called manually from hardcopy produced by the genotyper (Applied Biosystems) program.
Survival Analysis.
Kaplan–Meier survival functions were used so as to exclude the few mice that suffered morbidity due to nonplasmacytoma causes. The survival curves shown in Results are Kaplan–Meier plots. In the case of survival curves stratified by genotype, missing genotypes were imputed by using the software program F2 (http://biosun01.biostat.jhsph.edu/∼kbroman/), which incorporates the Lander–Green algorithm (16). The log-rank test (S-PLUS V5.1; www.splus.com) was used to test differences between survival curves.
Linkage Analysis.
All F2 mice were included in the linkage analyses. Linkage analysis for the X chromosome in male mice was performed by using nonparametric scans in Mapmaker/QTL.
Analysis of linkage of survival phenotypes to autosomal loci was performed by using the Mapmaker/SURVIVOR extension of mapmaker (17). This software computes LOD scores under the Cox proportional hazards model by using a variant of the expectation maximization algorithm using Monte Carlo simulation to make the computations tractable as described by Lipsitz et al. (18). The expectation–maximization algorithm incorporates all possible values of missing covariates according to the appropriate probability distributions based on the Lander–Green algorithm, both according to linked markers and maximum likelihood estimators of the coefficients for the baseline hazard functions. Mapmaker/SURVIVOR is available from M.J.D. at mjdaly@genome.wi.mit.edu. The test under the Cox proportional hazards model is equivalent to a Cox likelihood ratio test with full data. As such, the genome-wide threshold for significance in an F2 murine cross of 4.3 (19) is appropriate. Permutation testing using the data presented here confirmed this figure.
The survival differences due to the linked loci were graphed after completion of genotypes at markers using the Lander–Green algorithm as coded in the F2 program. Survival curves stratified by genotype were plotted by using s-plus. By way of comparison with the LOD scores, log-rank tests were performed at the linked loci by using s-plus.
Binary regression trees using data bagging and importance statistics were used to aid investigation of locus–locus interactions. Potential interactions were investigated further by using Cox likelihood ratio tests. The likelihood due to the loci acting alone was subtracted from the likelihood under a model including the putative locus–locus interaction and the effects of the individual loci. P values were computed from the likelihoods due to the locus–locus interactions under the χ2 distribution with 4 degrees of freedom.
Results
Plasmacytoma-Related Morbidity.
In the present study, >95% of the EμSV-v-abl transgenic mice that became ill by 1 year of age were found to harbor a tumor. All of these tumors were confirmed as plasmacytomas on the basis of their histologic appearance, as described (20). In addition, approximately 90% of the plasmacytoma-bearing mice had clonal paraproteins in their serum. Table 1 shows the frequency of pathologic findings in mice where there was gross or microscopic evidence of tumor at autopsy. The most common findings on gross examination were abdominal tumors, in particular tumors of the Peyer's patches, tumors arising from the mesenteric lymph nodes, and cecal tumors. Some instances of rapid-onset illness were observed, especially intussusception, intestinal hemorrhage, and small bowel obstruction, secondary to Peyer's patch and cecal tumors. Some mice had mediastinal tumors, sometimes causing respiratory distress; most had splenomegaly. Some mice were killed with hind limb paresis or paralysis, caused by plasmacytoma invading the spinal canal and compressing the spinal cord or the cauda equina.
Table 1.
Autopsy findings in transgene-carrying BCF2 animals
| Phenotype | Males and females | Males | Females |
|---|---|---|---|
| Peyer's patch tumors | 63.9 | 66.5 | 60 |
| Cecal tumor | 1.2 | 0.4 | 2.2 |
| Intussusception | 15.2 | 17.9 | 10.9 |
| Intestinal hemorrhage | 9.8 | 11.3 | 7.7 |
| Small bowel obstruction | 2.3 | 2.5 | 2.0 |
| Mesentery-associated tumor mass | 88.2 | 86.6 | 88.2 |
| Hind limb paralysis | 6.9 | 5.3 | 9.2 |
| Mediastinal tumor | 9.5 | 11.6 | 6.5 |
| Splenomegaly | 74.1 | 73.3 | 75.4 |
| Peripheral lymphadenopathy | 3.3 | 2.9 | 4.3 |
Numbers represent the percentage of tumor-bearing mice showing the given feature.
Plasmacytoma Susceptibility in BALB/c and C57BL/6 Mice and Genetic Crosses.
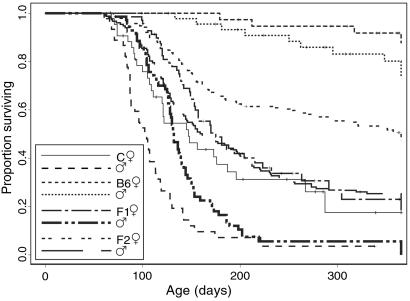
Fig. 1 shows Kaplan–Meier survival curves for inbred and hybrid v-abl 40-transgenic mice. Consistent with our previous results (8), 4.6% of the C.abl40 mice died before 10 weeks of age, a further 51.8% died between 10 and 20 weeks, and 89.2% failed to survive healthy to 1 year. Thus, these mice were markedly more susceptible than the B6.abl40 mice, of which 1.2% became ill with plasmacytoma before 20 weeks and 19.7% by 1 year of age.
Fig 1.
Comparison of survival of transgenic males and females of C.abl40 (BALB/c background, 88 mice), B6.abl40 (C57BL/6 background, 88 mice), 142 CBF1, and the first cohort of 332 CBF2 mice.
Tumor development in the F1 cohort of mice was slightly delayed compared with the C.abl40 mice (Fig. 1). In this cohort, 0.7% were killed with plasmacytoma before 10 weeks, a further 23.1% were killed between 10 and 20 weeks of age, and 92.7% were killed by 1 year. The F2 mice were bred in two cohorts separated by 1 year. Both had survival rates intermediate between those of the two parental strains. In the first cohort, 0.9% died before 10 weeks of age, 29% died between 10 and 20 weeks, and 61% died by 1 year (Fig. 1). The survival of animals in the second cohort is discussed below.
Susceptibility Differences Are Not Related to Transgene Expression.
Northern blot hybridization was performed to compare v-abl 40 transgene expression in the two parental mouse lines. Femoral bone marrow transgene expression in B6.abl40 and C.abl40 was compared. Nontransgenic C57BL/6 and BALB/c mice were used as controls. Expression of the v-abl 40 transgene in this primary lymphoid tissue was equivalent on C57BL/6 and BALB/c backgrounds (data not shown).
Sex and Parental Origin of Transgene Influence Susceptibility.
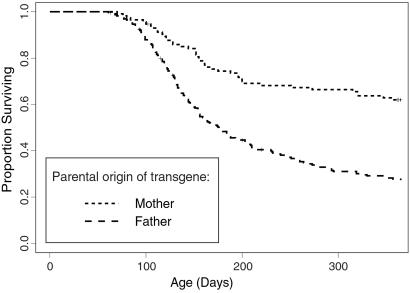
Susceptibility to the oncogenic effects of the transgene was influenced by the sex of the particular mouse, and also by the sex of the parent transmitting the transgene. Males were more susceptible than females (P = 2.4 × 10−4, 0.136, 7 × 10−7, and 7.9 × 10−15 for sex differences within C.abl40, B6.abl40, F1, and F2 generations, respectively). Fig. 2 presents a graphical comparison of the sex and strain differences. F2 generation mice inheriting the transgene from their fathers were more susceptible than mice inheriting the transgene from their mothers (P < 10−15). This presumably reflects some form of genomic imprinting.
Fig 2.
Comparison of survival among 332 abl40-transgenic CBF2 generation mice in the first cohort stratified by parental origin of the transgene.
Rearing in an SPF Facility Decreases Susceptibility.
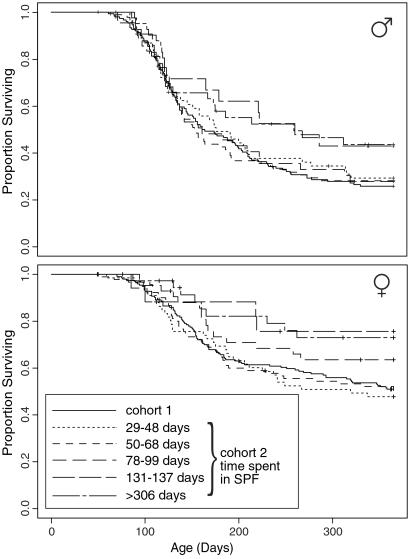
The first cohort of 332 F2 mice was bred and maintained in our open access mouse facility. Because of space constraints the second cohort of 509 F2 mice was reared in an SPF facility. The majority were gradually transferred to the open access facility. The approximate dates when transfers occurred were established retrospectively, and five cohorts were identifiable, comprising 126 mice transferred at 29–48 days, 182 at 50–68 days, 85 at 78–99 days, and 39 at 131–137 days of age, and the remaining 77 that spent the entire 365 days of the experiment in SPF. The survival curves of these cohorts of mice are compared in Fig. 3.
Fig 3.
Comparison of survival of subgroups of the second cohort of 509 abl40-transgenic CBF2 mice stratified by time spent in SPF conditions and the first cohort of F2 mice. (Upper) Survival curves for the male mice. (Lower) Survival curves for the female mice.
The survival curves of the males fell into two groups. Those of the three cohorts transferred between days 29 and 99 were almost coincident. By comparison, the mice transferred at 131–137 days of age and those that were never transferred also had almost coincident survival curves, and were much less susceptible to plasmacytoma development. The survival curves diverge minimally for mice transferred before day 126. The majority of the difference in tumor susceptibility is exhibited after this time point.
The females exhibited a similar pattern. The most susceptible were those transferred between days 29 and 68. The animals transferred at 78–99 days had intermediate susceptibility. Least susceptible were those transferred at 131–137 days or never transferred.
Linkage Results for Loci with Independent Effects.
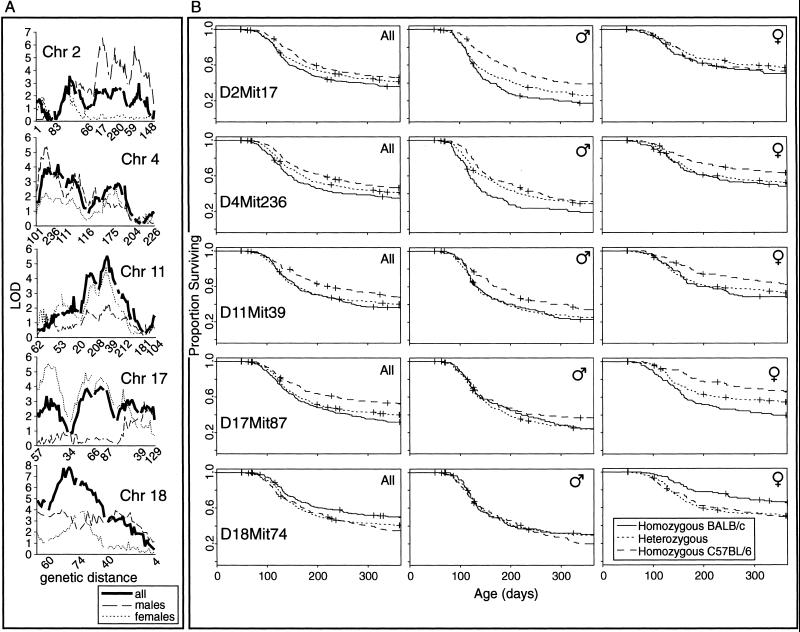
The use of Mapmaker/SURVIVOR, a computational algorithm for detecting linkage to survival phenotypes, is described here. Fig. 4 and Table 2 summarize the single-locus linkage results. The linkage scores at the most significantly linked loci are shown. Significance was accepted as a LOD score greater than 4.3. The LOD and χ2 scores at markers are shown to enable comparison of Mapmaker/SURVIVOR with the log-rank test. Phenotypic differences between the sexes suggested that stratification by sex in the analyses might be informative, so linkage analysis was performed on the male and female mice separately as well as together.
Fig 4.
Linked autosomal loci and associated survival phenotypes in 835 abl40-transgenic CBF2 mice. (A) Chromosomal scans for the five linked autosomes (centromere at left). LOD scores for all of the mice together as well as after stratification by sex are shown. Selected Mit markers are indicated along each chromosome. (B) Survival curves at the most tightly linked markers. Each graph shows three survival curves, one for each of the possible genotypes. Three graphs are shown for each marker, taking all of the mice together, and each sex separately.
Table 2.
Loci most tightly linked to plasmacytoma-free survival using single-locus statistics
| Chromosome
|
Interval containing peak
|
LOD at peak | Closest marker
|
Position, cM
|
LOD at marker | χ2 at marker | Susceptible allele
|
Genetics
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All | Males | Females | All | Males | Females | All | Males | Females | ||||||
| 2 | D2Mit17 | 2.1 | 6.1 | 0.1 | D2Mit17 | 56.8 | 2.4 | 5.6 | 0.0 | 6.8 | 16.1 | 1.0 | BALB/c | Additive; Male only |
| 4 | D4Mit101–D4Mit236 | 3.9 | 5.2 | 2.3 | D4Mit236 | 16.4 | 3.6 | 3.2 | 1.6 | 10.0 | 12.3 | 5.9 | BALB/c | Additive |
| 11 | D11Mit208–D11Mit39 | 5.5 | 2.2 | 4.3 | D11Mit39 | 44.8 | 3.2 | 1.4 | 2.4 | 5.8 | 5.9 | 1.8 | BALB/c | Dominant |
| 17 | D17Mit57–D17Mit34 | 2.6 | 0.4 | 5.6 | D17Mit57 | 1.1 | 2.2 | 0.2 | 3.8 | 3.8 | 1.3 | 6.4 | BALB/c | Additive; See text concerning females |
| D17Mit66–D17Mit87 | 4.0 | 0.6 | 4.6 | D17Mit87 | 29.5 | 4.0 | 0.0 | 4.4 | 19.9 | 5.0 | 19.6 | BALB/c | ||
| 18 | D18Mit60–D18Mit74 | 7.8 | 4.0 | 3.9 | D18Mit74 | 16.4 | 6.0 | 2.2 | 3.9 | 3.5 | 0.9 | 2.4 | C57BL/6 | Additive |
Maximal LOD scores are given, along with the LOD score at the most tightly linked marker used. For comparison, the results of log-rank tests at these markers after imputation of missing genotypes are provided. Results are given for all the mice, as well as results after stratification on the basis of sex. Comments are made as to the nature of the genetic effects of the alleles at these loci.
Single-locus analysis revealed a number of interesting features. In particular, survival is modified by different loci in males and females. Survival of male mice is affected most strongly by the allelotype at D2Mit17, which has a negligible effect on the survival of females. D17Mit87 and D18Mit74 are important modifiers of survival in females. The allelic complement at D4Mit111 and D11Mit39 affects survival in both male and female mice.
DXMit55 may be linked to survival. Unfortunately it is impossible to examine the effects of this locus on both male and female mice in the same analysis. Comparatively modest linkage scores were observed for both males and females (Z = 1.3 and LOD = 2.3, respectively). In both males and females the BALB/c alleles of the X chromosome locus were associated with relative susceptibility.
Possible Locus–Locus Interaction.
Locus–locus interactions were investigated by using binary regression trees. Among the female mice, two pairs of loci were identified that appeared to interact. One locus seems significant. Table 3 shows the two interactions along with the P value associated with the interaction. The Cox proportional hazards model was used to determine the log-likelihood caused by the loci and any interaction between them, as well as the log-likelihood caused by either of the loci with independent effects acting alone. The latter was subtracted from the former to obtain a log-likelihood caused by the interaction alone, and it was converted into a P value. The D17Mit87–D2Mit148 interaction in females explains the survival substantially better than do the underlying loci, whereas the P value of 0.09 shows that the same is not true for males.
Table 3.
The significance levels of the two interactions suggested by binary regression-tree analysis to be the most important
| Locus pair
|
Female | Male | ||||
|---|---|---|---|---|---|---|
| Log likelihood caused by locus pair |
P
|
Log likelihood caused by locus pair |
P
|
|||
| Including interaction | Not including interaction | Including interaction | Not including interaction | |||
| D17Mit87–D2Mit148 | 37.3 | 21.5 | 0.003 | 22.2 | 14.2 | 0.09 |
| D17Mit87–D11Mit212 | 35.8 | 33.3 | 0.650 | 11.6 | 10.5 | 0.90 |
The likelihoods for models including the two loci with or without interactions are shown. The P values were calculated from the difference between the log likelihoods.
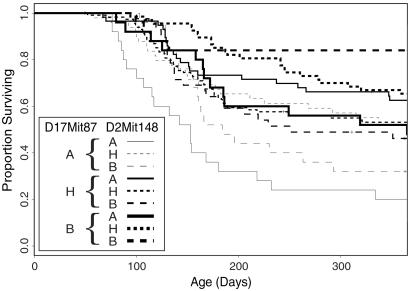
Graphs of survival stratified by both loci in a pair simultaneously demonstrate the manner in which the loci interact. The interaction between D2Mit148 and D17Mit87 in females is complex (Fig. 5). When the genotype at D17Mit87 is either homozygous BALB/c or homozygous C57BL/6, then BALB/c homozygosity at D2Mit148 confers relative susceptibility. The effect of BALB/c D2Mit148 alleles on the phenotype is roughly additive in the case of C57BL/6 homozygosity at D17Mit87. However, where D17Mit87 is homozygous for BALB/c alleles, although BALB/c homozygotes at D2Mit148 are more susceptible than C57BL/6 homozygotes at D2Mit148, the most resistant mice are heterozygous at D2Mit148. Among mice heterozygous at D17Mit87, BALB/c homozygotes at D2Mit148 are more resistant than C57BL/6 homozygotes, and heterozygotes have intermediate susceptibility.
Fig 5.
Survival of female abl40-transgenic CBF2 animals after stratification at both D17Mit87 and D2Mit148 to show the effects of the interaction between these two markers. “A” represents BALB/c homozygosity, “H” represents heterozygosity, and “B” represents C57BL/6 homozygosity.
D2Mit148 did not exert a detectable effect in single-locus tests. The complex nature of the interaction between the two loci explains why D2Mit148 was not individually linked to susceptibility; its effect appears to depend on the alleles present at the locus linked to D17Mit87. The putative interaction between these loci may be sufficient to explain the susceptibility difference between female B6.abl40 and C.abl40 mice. The 365-day survival rate in mice homozygous for C57BL/6 alleles at both loci was 84%, and for mice homozygous for BALB/c alleles at both loci it was 20%. This is essentially the same difference in survival as seen between v-abl transgenic female C57BL/6 and BALB/c mice.
Transgene Mapping.
Analysis of the genotypic data shows that the transgene is tightly linked to an SJL allele at D14Mit133, and less tightly linked to C57BL/6 alleles at flanking markers. Thus, the original transgene insertion in the founder (C57BL/6 × SJL/J)F2 embryo was into SJL chromatin, a small segment of which has been retained by selection through the back-crossing onto the C57BL/6 background.
Discussion
A difference in the incidence of plasmacytoma induced by a v-abl transgene in two inbred strains of mice, BALB/c and C57BL/6, has been observed. The present, more extensive study confirms the much higher susceptibility of BALB/c mice. It makes the additional finding that in both strains males are more susceptible than females. This finding echoes the sex difference in the incidence of the analogous tumor, multiple myeloma in humans. In the well-studied alternative model of mouse plasmacytoma induced by chronic peritoneal irritation, BALB/c mice are also highly susceptible (21), but a sex difference is not found consistently in that system (22, 23).
Again, unlike the peritoneal irritant-induced tumors (21), first-generation v-abl-transgenic hybrids between BALB/c and C57BL/6 strains proved to be almost as prone to plasmacytoma development as inbred BALB/c mice. This near dominance of high susceptibility, plus the large difference in tumor incidence between the two inbred strains, seemed to indicate a promising subject for a tumor modifier mapping study. However, a genome-wide scan of an initial group of 332 (BALB/c × C57BL/6)F2 mice for linkage of microsatellite-based alleles with survival time revealed a high level of genetic complexity. This finding necessitated expansion of the experiment into one of the largest tumor modifier mapping studies reported to date; we tracked the health of 842 v-abl-transgenic F2 mice, checked all that became ill for evidence of plasmacytoma, and determined the genotype of almost every mouse at 191 markers. The study also prompted our creation and use of the Mapmaker/SURVIVOR linkage algorithm.
A further complexity was added by the observation that a subgroup of F2 mice that was housed in an SPF facility for 4 months or more showed a lower incidence of plasmacytoma than the majority of F2 mice, which were housed in a conventional facility. This phenomenon operates even more strikingly in peritoneal irritant-induced plasmacytoma, with germ-free (24) and SPF (25) BALB/c mice being refractory. Unfortunately, the nature of the relevant environmental difference between the two facilities in the present experiment is unclear, but it is likely to be microbial. This finding offers another possible point of confluence between experimental mouse plasmacytoma and human multiple myeloma. There have been indications that chronic inflammatory disease (26), but probably not exogenous antigen exposure (27), alters a person's risk of developing myeloma.
Analysis for single-locus linkage to survival time in the v-abl-transgenic F2 mice revealed one or more significantly influential regions on each of five different chromosomes. Some of these loci were sex-specific in their action. One or more loci on chromosome 2, for example, modified susceptibility in males but not females, whereas loci on chromosomes 17 and 18 affected tumorigenesis predominantly in females. With most of the linked loci, the BALB/c allelotype conferred high susceptibility, but for the major locus on chromosome 18, C57BL/6 haplotypes were associated with a higher incidence of plasmacytoma.
The linkage scores obtained from Mapmaker/SURVIVOR compare favorably with those obtained by genotype imputation and subsequent log-rank tests at loci. This finding demonstrates the advantage of using this software, which provides linkage analysis for survival phenotypes in a package combining, in the Mapmaker tradition, interval mapping and an expectation–maximization style algorithm for imputing missing covariates. This software will prove useful in the analysis of all forms of survival data, or other data where time of onset to a particular phenotype is thought to be important.
Tests for epistasis revealed two possibly interacting loci linked to D2Mit148 and D17Mit87, which could explain most of the genetically determined difference in susceptibility of female mice of the two strains. The P value of 0.003 is suggestive of the validity of this interaction, without establishing formal significance on a genome-wide level. The nature of the interaction appears complex.
The biological significance of the single-locus effects could be assessed by developing mouse strains congenic for each potentially interesting locus and measuring the incidence of plasmacytoma in them. Likewise, it may be possible to demonstrate the epistatic interaction using doubly congenic mice.
Estimates of the heritability of tumor susceptibility imply the existence of more tumor modifier genes than are currently known (1). That these genes are yet unidentified suggests that they are involved in complex interactions, individually have small effects, or both. It is noteworthy that one of the most successful studies of this kind, the mapping of the Modifier of Min-1 locus (28), has led to the implication of the secretory phospholipase A2 gene (29, 30), which would not have been considered as a tumor modifier from the previous biological understanding. Identification of the genes underlying the loci identified here should cast light on susceptibility to onset and development of multiple myeloma, including the effects of sex, and thereby on immunologic physiology and the biochemical systems of relevance to tumor susceptibility in general.
Acknowledgments
We thank A. Mifsud, C. Tilbrook, E. Shomali, K. Gray, H. Millar, D. Postma, and T. Gibbs for their excellent care of the mice; and Dr. R. Thomson, Dr. M. L. Bath, the Australian Genome Research Facility, and the histology department of the Walter and Eliza Hall Institute for technical assistance. This work was funded by The Wellcome Trust (London) and the National Health and Medical Research Council, Australia.
Abbreviations
LOD, logarithm of odds
SPF, specific pathogen free
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Lichtenstein P., Holm, N. V., Verkasalo, P. K., Iliadou, A., Kaprio, J., Koskenvuo, M., Pukkala, E., Skytthe, A. & Hemminki, K. (2000) N. Engl. J. Med. 343, 78-85. [DOI] [PubMed] [Google Scholar]
- 2.Hopper J. L., Southey, M. C., Dite, G. S., Jolley, D. J., Giles, G. G., McCredie, M. R., Easton, D. F. & Venter, D. J. (1999) Cancer Epidemiol. Biomarkers Prev. 8, 741-747. [PubMed] [Google Scholar]
- 3.Zhang S., Ramsay, E. S. & Mock, B. A. (1998) Proc. Natl. Acad. Sci. USA 95, 2429-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang S. & Mock, B. A. (1999) Curr. Top. Microbiol. Immunol. 246, 363-367. [DOI] [PubMed] [Google Scholar]
- 5.Mock B. A., Hartley, J., Le Tissier, P., Wax, J. S. & Potter, M. (1997) Blood 90, 4092-4098. [PubMed] [Google Scholar]
- 6.Ries L. A. G., Eisner, M. P., Kosary, C. L., Hankey, B. F., Miller, B. A., Clegg, L. & Edwards, B. K., (2002) SEER Cancer Statistics Review, 1973–1999 (Natl. Cancer Inst., Bethesda), http://seer.cancer.gov/csr/1973_1999.
- 7.Rosenbaum H., Harris, A. W., Bath, M. L., McNeall, J., Webb, E., Adams, J. M. & Cory, S. (1990) EMBO J. 9, 897-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris A. W., Strasser, A., Bath, M. L., Elefanty, A. G. & Cory, S. (1997) Curr. Top. Microbiol. Immunol. 224, 221-230. [DOI] [PubMed] [Google Scholar]
- 9.Kruglyak L. & Lander, E. S. (1995) Genetics 139, 1421-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mock B. A., Krall, M. M. & Dosik, J. K. (1993) Proc. Natl. Acad. Sci. USA 90, 9499-9503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Potter M., Mushinski, E. B., Wax, J. S., Hartley, J. & Mock, B. A. (1994) Cancer Res. 54, 969-975. [PubMed] [Google Scholar]
- 12.Piechaczyk M., Blanchard, J. M., Marty, L., Dani, C., Panabieres, F., El Sabouty, S., Fort, P. & Jeanteur, P. (1984) Nucleic Acids Res. 12, 6951-6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Copeland N. G., Gilbert, D. J., Jenkins, N. A., Nadeau, J. H., Eppig, J. T., Maltais, L. J., Miller, J. C., Dietrich, W. F., Steen, R. G. & Lincoln, S. E. (1993) Science 262, 67-82. [DOI] [PubMed] [Google Scholar]
- 14.Dietrich W. F., Miller, J. C., Steen, R. G., Merchant, M., Damron, D., Nahf, R., Gross, A., Joyce, D. C., Wessel, M. & Dredge, R. D. (1994) Nat. Genet. 7, 220-245. [DOI] [PubMed] [Google Scholar]
- 15.Dietrich W. F., Miller, J., Steen, R., Merchant, M. A., Damron-Boles, D., Husain, Z., Dredge, R., Daly, M. J., Ingalls, K. A. & O'Connor, T. J. (1996) Nature (London) 380, 149-152. [DOI] [PubMed] [Google Scholar]
- 16.Lander E. S., Green, P., Abrahamson, J., Barlow, A., Daly, M. J., Lincoln, S. E. & Newburg, L. (1987) Genomics 1, 174-181. [DOI] [PubMed] [Google Scholar]
- 17.Lander E. S. & Botstein, D. (1989) Genetics 121, 185-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipsitz S. R. & Ibrahim, J. G. (1998) Biometrics 54, 1002-1013. [PubMed] [Google Scholar]
- 19.Lander E. S. & Schork, N. J. (1994) Science 265, 2037-2048. [DOI] [PubMed] [Google Scholar]
- 20.Harris A. W., Bath, M. L., Rosenbaum, H., McNeall, J., Adams, J. M. & Cory, S. (1990) Curr. Top. Microbiol. Immunol. 166, 165-173. [DOI] [PubMed] [Google Scholar]
- 21.Potter M., Pumphrey, J. G. & Bailey, D. W. (1975) J. Natl. Cancer Inst. 54, 1413-1417. [DOI] [PubMed] [Google Scholar]
- 22.Takakura K., Yamada, H., Weber, A. H. & Hollander, V. P. (1967) Cancer Res. 27, 932-937. [PubMed] [Google Scholar]
- 23.Potter M. & Wax, J. (1983) J. Natl. Cancer Inst. 71, 391-395. [PubMed] [Google Scholar]
- 24.McIntire K. R. & Princler, G. L. (1969) Immunology 17, 481-487. [PMC free article] [PubMed] [Google Scholar]
- 25.Byrd L. G., McDonald, A. H., Gold, L. G. & Potter, M. (1991) J. Immunol. 147, 3632-3637. [PubMed] [Google Scholar]
- 26.O'Byrne K. J. & Dalgleish, A. G. (2001) Br. J. Cancer 85, 473-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis D. R., Pottern, L. M., Brown, L. M., Silverman, D. T., Hayes, R. B., Schoenberg, J. B., Greenberg, R. S., Swanson, G. M., Schwartz, A. G., Liff, J. M., et al. (1994) Cancer Causes Control 5, 529-539. [DOI] [PubMed] [Google Scholar]
- 28.Dietrich W. F., Lander, E. S., Smith, J. S., Moser, A. R., Gould, K. A., Luongo, C., Borenstein, N. & Dove, W. (1993) Cell 75, 631-639. [DOI] [PubMed] [Google Scholar]
- 29.Cormier R. T., Hong, K. H., Halberg, R. B., Hawkins, T. L., Richardson, P., Mulherkar, R., Dove, W. F. & Lander, E. S. (1997) Nat. Genet. 17, 88-91. [DOI] [PubMed] [Google Scholar]
- 30.Cormier R. T., Bilger, A., Lillich, A. J., Halberg, R. B., Hong, K. H., Gould, K. A., Borenstein, N., Lander, E. S. & Dove, W. F. (2000) Oncogene 19, 3182-3192. [DOI] [PubMed] [Google Scholar]