Abstract
The tumor suppressor function of p53 has been attributed to its ability to regulate apoptosis and the cell cycle. In mammals, DNA damage, aberrant growth signals, chemotherapeutic agents, and UV irradiation activate p53, a process that is regulated by several posttranslational modifications. In Drosophila melanogaster, however, the regulation modes of p53 are still unknown. Overexpression of D. melanogaster p53 (Dmp53) in the eye induced apoptosis, resulting in a small eye phenotype. This phenotype was markedly enhanced by coexpression with D. melanogaster Chk2 (DmChk2) and was almost fully rescued by coexpression with a dominant-negative (DN), kinase-dead form of DmChk2. DN DmChk2 also inhibited Dmp53-mediated apoptosis in response to DNA damage, whereas overexpression of Grapes (Grp), the Drosophila Chk1-homolog, and its DN mutant had no effect on Dmp53-induced phenotypes. DmChk2 also activated the Dmp53 transactivation activity in cultured cells. Mutagenesis of Dmp53 amino terminal Ser residues revealed that Ser-4 is critical for its responsiveness toward DmChk2. DmChk2 activates the apoptotic activity of Dmp53 and Ser-4 is required for this effect. Contrary to results in mammals, Grapes, the Drosophila Chk1-homolog, is not involved in regulating Dmp53. Chk2 may be the ancestral regulator of p53 function.
Various forms of cellular stress such as DNA damage or ionizing irradiation lead to activation and stabilization of the p53 tumor suppressor protein and to growth arrest and apoptosis (1, 2). Chk2, the mammalian homolog of the Saccharomyces cerevisiae Rad 53 and the Schizosaccharomyces pombe Cds1 checkpoint genes (3, 4), regulate p53 function in mammals in response to DNA damage (5). Chk2 is a protein kinase that acts downstream of the ataxia telangiectasia mutated (ATM) kinase and may induce cell cycle arrest (6, 7). Loss of Chk2 in thymocytes results in failure to increase intracellular p53 levels in response to DNA damage, causing a defect in p53-mediated apoptosis (8). Chk1, an evolutionarily conserved protein kinase, implicated in cell cycle checkpoint control in lower eukaryotes (9–11), also has been suggested to play a role in p53 regulation (3, 4). Chk1 also can phosphorylate p53, probably at the same sites as Chk2 (12). Currently, the relative significance of p53 phosphorylation by Chk2 and/or Chk1 in the process of p53 activation is unclear. Several components involved in cell cycle checkpoint control pathways are conserved in Drosophila. Drosophila homologs of the ATM/ATR (mei-41), Chk1 (grapes), and Chk2 (loki) kinases have been identified (13–15). Mei-41 mutant cells are sensitive to ionizing radiation, display high levels of mitotic chromosome instability, and do not arrest upon radiation treatment (13). Grapes (Grp) has been shown to be involved in a developmentally regulated DNA replication/damage checkpoint operating during the late syncytial divisions (14). The Drosophila maternal nuclear kinase (DMNK) protein is the homolog of the human Chk2 protein [referred to as Drosophila melanogaster Chk2 (DmChk2)] and is highly expressed in Drosophila ovaries and functions in meiosis (15). The role of DmChk2 or Grp in the regulation of D. melanogaster p53 (Dmp53) is unknown. It is also not clear, as to whether DmChk2 functions in a cell cycle checkpoint pathway in vivo. Data from Caenorhabditis elegans suggest that Chk2 mutants are defective in meiosis but retain a DNA damage checkpoint in response to replication inhibition and ionizing radiation (16, 17). Cloning and characterization of Dmp53 was recently described showing an essential role for Dmp53 in radiation-induced apoptosis (18–20). It was found that overexpressing of Dmp53 in the fly eye resulted in massive apoptosis and a small eye phenotype (18–20). We have now built on these results by using genetic epistasis experiments and demonstrate an essential role for DmChk2 in regulation of Dmp53. The amino-terminal Ser at position 4 of the Dmp53 molecule confers responsiveness toward DmChk2. Interestingly, Grp is not involved in the regulation of Dmp53 in this system.
Materials and Methods
Cloning of Dmp53 cDNA.
A tblastn search of the raw data from the Celera Genomics D. melanogaster Genome Database identified a genomic fragment spanning the region encoding amino acids 97–385 of human p53 (i.e., SwissProt accession no. P04637) with P = 5e-19. Oligonucleotide primers flanking the putative Dmp53 ORF were designed (sense primer 5′-CTA AGA TGT ATA TAT CAC AGC CAA TGT CGT GG-3′ and antisense primer 5′-GCC ATC GAA CAT GCC AAA AGG GGA ATA TCA CC-3′) and used to amplify a 1.32-kb product from D. melanogaster 18-h embryo QUICK-Clone cDNA (CLONTECH).
Generation of Anti-Drosophila- and Anti-Human p53 Abs.
mAbs were raised according to the method of Kohler and Milstein (21). Individual clones were subsequently screened by Western blot and immunofluorescence.
In Vitro Kinase Assay.
293T cells transfected for 48 h, with 4 μg of plasmids expressing Myc/His-DmGrp or Myc/His-DmChk2, and 4 or 8 μg or the respective mutants, Myc/His-DmGrp-D130A or Myc/His-DmChk2-D303A, were lysed in CHAPS (3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate) buffer and immunoprecipitated by using an anti-Myc mAb and protein A agarose beads. Beads were washed with kinase buffer (50 mM Tris⋅HCl, pH 7.5/0.1 mM EGTA/1.5 mM DTT/1 μg/ml BSA) and incubated with 32PγATP and the synthetic Chk1/Chk2 substrate peptide (KKKVSRSGLYRSPSMPENLNRRR) (Upstate Biotechnology) for 10 min at 30°C (22, 23). Peptides were resolved by SDS/PAGE and visualized by autoradiography.
Immunohistochemistry and Analysis of Flies.
p53 was detected in fixed eye discs using the above-described Abs. In brief, third-instar larval heads were dissected and fixed in 2% paraformaldehyde and stained overnight at 4°C with the respective Ab [1:10 (Drosophila) or 1:50 (human) in PBS plus 0.1% Triton X]. For BrdUrd labeling, third-instar larval heads were dissected in PBS and incubated for 1 h in M3 insect media (Sigma) containing 100 μg/ml BrdUrd (Sigma) and then fixed in 2% paraformaldehyde. Apoptotic cells in the developing eyes were visualized with the vital dye acridine orange by using standard methods. Scanning electron microscopy was performed by using standard techniques with an Olympus electron microscope.
Western Blotting of Larval Extracts.
Thirty third-instar larvae were lysed with a Dounce homogenizer in 500 μl of lysis buffer (5% SDS/20 mM Hepes, pH 7.5/5 mM EGTA/5 mM EDTA). Extracts were boiled and protein samples were resolved by SDS/PAGE and transferred to PVDF [poly(vinylidene difluoride)] membrane. Blots were probed with mouse anti-Chk1 Ab (Santa Cruz Biotechnology) at a dilution of 1:500 and rabbit anti-actin Ab (Sigma) at a dilution of 1:200. Secondary horseradish peroxidase-conjugated Abs were used at a dilution of 1:1,000.
Tissue Culture Experiments.
Drosophila S2 cells (3 × 106) were transfected by using 20 μg of total DNA mixed with CellFectin reagent (GIBCO) in six-well plates according to methods described (18). For experiments shown in Fig. 5, cells were induced with CuSO4 24 h after transfection and were harvested 18 h after induction. Chloramphenicol acetyltransferase (CAT) activity was normalized to β-galactosidase activity.
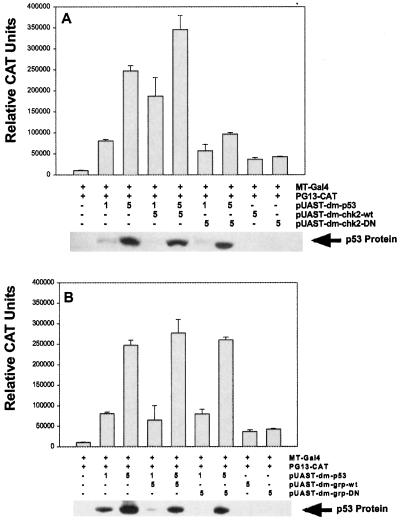
Fig 5.
Chk2 regulates p53 transactivation in cultured cells. Drosophila S2 cells were transfected with 1 or 5 μg of wild-type Dmp53 to test for transcriptional activation from PG13, a promoter containing wild-type binding sites for human p53. (A) Cells were cotransfected with 5 μg of wild-type DmChk2 or a DN, kinase-dead form of DmChk2. (B) Cells were cotransfected with 5 μg of either wild-type Grp or a DN, kinase-dead form of Grp. Error bars indicate the SEM from three independent experiments. (A Lower and B Lower) Expression of Dmp53 protein in the respective lysates.
FACS Analysis.
Eye imaginal discs (30–50) were dissociated by incubation with trypsin and analyzed by flow cytometry as described (18). The cell suspension was analyzed by using a Becton Dickinson FACS Vantage flow cytometer, and the data were analyzed with cell quest software (Becton Dickinson).
Fly Genetics.
The UAS/Gal4 system (25) was used to drive expression of human p53, Dmp53, DmChk2, DmChk2-D303A, Grp, and Grp-D130A. GMR-Gal4 (glass multimer reporter) transgenic flies were used in all experiments (21). The following genotypes were used in the experiments: GMR-Gal4,UAS-human-p53/CyO; GMR-Gal4,UAS-Dmp53/CyO; UAS-DmChk2/TM3; UAS-Grp/TM3; UAS-DmChk2-D303A/TM3 [DmChk2-dominant negative (DN)]; and UAS-Grp-D130A/TM3(Grp-DN). Flies overexpressing a DN form of Dmp53 (D259H) have been described (14). Flies were kept at 25°C during all experiments. All experiments were performed with at least two different transgenic lines.
Results
Characterization of p53 Transgenic Flies.
Overexpression of human p53 in the Drosophila eye resulted in a striking reduction in eye size and a disruption of the ommatidia structure (Fig. 1C). Ommatidia were fused and exhibited a complete loss of bristles. Overexpression of Dmp53 produced a less severe phenotype, resulting in a reduced eye size with partial fusion of the ommatidia and some remaining bristles (Fig. 1B). Consistent with the known activity of the GMR promoter, the expression of both proteins was detected in the eye imaginal disc posterior to the morphogenetic furrow (Fig. 1 E and F).
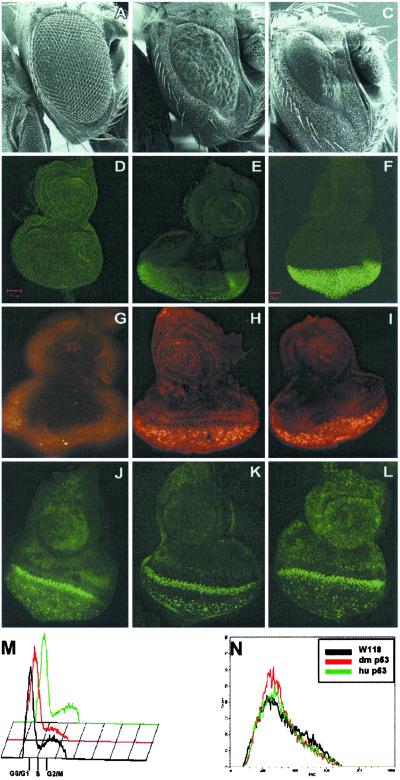
Fig 1.
Effects of hp53 or Dmp53 overexpression in the fly eye. (A–C) Scanning electron micrographs of eyes from GMR-Gal4 flies crossed to a wild-type fly (A), UAS-Dmp53 fly (B), or UAS-hp53 fly (C). (Magnification: ×220.) (D–F) Expression of p53 in eye discs. Immunohistochemical detection of Dmp53 in wild-type (D), and UAS-Dmp53 (E) or hp53 in UAS-hp53 (F) flies crossed to GMR-Gal4. (G–I) Distribution of dying cells in eye discs by using acridine orange. Wild-type larva (G), UAS-Dmp53 larva (H), or hp53 larva (I) crossed to GMR-Gal4 flies. (J–L) Cellular proliferation shown by BrdUrd labeling. Wild-type flies (J), Dmp53 overexpressing flies (K), and hp53 overexpressing flies (L). (M and N) Cell cycle analysis in single cell suspensions dissected from eye discs. (Magnification: ×400.) (M) Histogram displaying DNA content and cell numbers from single cell suspensions from eye discs dissected from wild-type (black), Dmp53 (red), and human p53 (green) overexpressing flies. (N) Forward side scatter displaying cell size and cell number.
Expression of Drosophila or human p53 resulted in extensive apoptosis in the developing eye, as judged by acridine orange staining (Fig. 1 H and I). The S-phase band posterior to the morphogenetic furrow, as visualized by BrdUrd immunohistochemistry, was present in both hp53 and Dmp53 transgenic flies (Fig. 1 K and L). No considerable differences in cell cycle distribution or cell size could be discerned when human p53 or Dmp53-overexpressing flies were compared (Fig. 1 M and N). These results show that overexpression of either human or Dmp53 resulted in apoptosis in the developing fly eye without detectable effects on cell cycle regulation.
DmChk2 Regulates the Function of Dmp53.
To study the interaction between Chk2 and p53, the cDNA encoding the Drosophila homolog of human Chk2 (DmChk2) was cloned. A kinase-dead form of DmChk2 was generated, in which the conserved aspartic acid at position 303 was mutated to Ala (DN-DmChk2). Mutation of this amino acid has been shown to function as a DN mutation (6).
DmChk2 and DN-DmChk2 were overexpressed in the Drosophila eye, under the control of the GMR promoter. No overt phenotype could be observed and no apoptotic cells were detected in the eye imaginal discs from third-instar larvae (Fig. 2 A and B and data not shown).
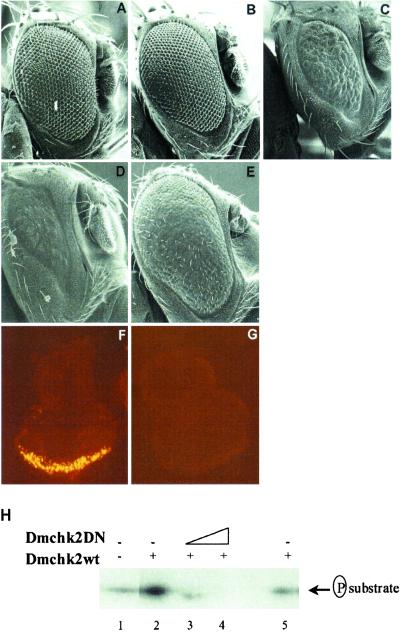
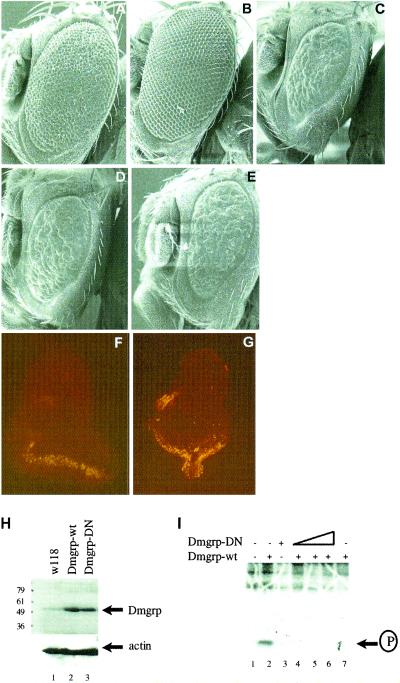
Fig 2.
Chk2 regulates p53 in Drosophila. (A–E) Scanning electron micrographs of eyes from flies overexpressing DmChk2 (A), DN-DmChk2 (B), Dmp53 (C), DmChk2 and Dmp53 (D), or DN-DmChk2 and Dmp53 (E), driven by the GMR-Gal4 promoter. (Magnification: ×220.) (F and G) Chk2-regulation of p53 induced apoptosis. Eye discs from a fly overexpressing DmChk2 and Dmp53 (F) or DN-DmChk2 and Dmp53 (G), driven by the GMR-Gal4 promoter, were stained with the vital dye acridine orange. (Magnification: ×400.) (H) DN-DmChk2 inhibits DmChk2 kinase activity. Myc/His-tagged DmChk2, alone (lane 2) or together with increasing amounts of DN-DmChk2 (lanes 3 and 4), immunoprecipitated from 293T cells, was incubated with synthetic Chk1/Chk2 substrate peptide and the resulting phosphorylation was assessed by autoradiography. As a control (lane 5), the anti-Myc/His Ab was omitted in the immunoprecipitation step.
To study the genetic interaction between Dmp53 and DmChk2, transgenic flies overexpressing Dmp53 (Fig. 2C) were crossed with transgenic flies overexpressing either wild-type or DN-DmChk2. Coexpression of wild-type DmChk2 and Dmp53 resulted in a considerably more severe phenotype with almost complete loss of the eye (Fig. 2D) compared to flies expressing Dmp53 alone (Fig. 2C). Excessive apoptosis was detected in eye imaginal discs from third-instar larvae coexpressing DmChk2 and Dmp53 (Fig. 2F). In contrast, coexpression of DN-DmChk2 with Dmp53 resulted in an almost complete rescue of the Dmp53-induced eye phenotype (Fig. 2E). Consistent with the observed eye morphology, no apoptosis could be found in eye imaginal discs in these animals (Fig. 2G). Analysis of Dmp53 protein levels by immunohistochemistry in flies coexpressing wild-type or DN-DmChk2 and Dmp53 revealed no difference compared to flies expressing Dmp53 alone, indicating that altered Dmp53 protein levels did not account for the observed changes in eye phenotype (data not shown).
A kinase assay was performed to confirm DmChk2 activity. DmChk2 phosphorylated a synthetic Chk1/Chk2 peptide substrate (Fig. 2H, lane 2). In the presence of increasing amounts of DN-DmChk2, the kinase activity of the wild-type DmChk2 was lost (Fig. 2H, lanes 3 and 4). These experiments show that the D303A mutant of DmChk2 functions as a true DN protein, inhibiting the kinase activity of the wild-type protein.
DN-p53 and Chk2 Both Protect from Irradiation-Induced Apoptosis in Drosophila.
Dmp53-mediated sensitivity to irradiation was investigated in wild-type and transgenic flies overexpressing a DN form of Dmp53 (Dmp53-D259H). The D259H point mutation corresponds to the human p53 mutational hotspot at position 273. In human p53, this amino acid directly contacts DNA and is required for DNA binding (26). Eye imaginal discs were dissected from third-instar larvae 4 h after γ-irradiation, and apoptotic cells were visualized with acridine orange. Wild-type, unirradiated eye discs did not show apoptotic cells (Fig. 3A). In contrast, irradiated wild-type eye discs exhibited a high number of apoptotic cells both in the antenna discs and in the anterior and posterior part of the eye imaginal discs (Fig. 3B). Irradiated eye discs from flies overexpressing Dmp53-D259H showed abundant apoptotic cells only anterior to the morphogenetic furrow, where Dmp53-D259H is not expressed. Posterior to the morphogenetic furrow, where Dmp53-D259H is expressed, few apoptotic cells were present (Fig. 3C). Our results show that Dmp53 is implicated in γ-irradiation-induced apoptosis in the Drosophila eye.
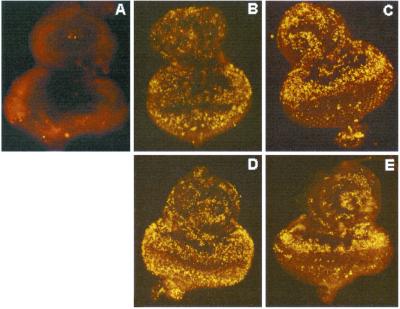
Fig 3.
DN forms of p53 and Chk2 prevent irradiation-induced apoptosis. Acridine orange-stained eye discs from transgenic flies treated without (A) or with (B–E) 40 Gy of γ-irradiation. Wild type, unirradiated (A), wild type (B), Dmp53–259H (C), DmChk2 (D), and DN-DmChk2 (E), driven by GMR-Gal4. (Magnifications: ×400.)
Irradiated eye imaginal discs overexpressing wild-type DmChk2 posterior to the morphogenetic furrow showed high numbers of acridine orange-positive apoptotic cells in this area (Fig. 3D). However, in DN-DmChk2-overexpressing eye discs, there was almost a complete absence of apoptotic cells, comparable to discs overexpressing the DN form of Dmp53 (Fig. 3E). These results demonstrate a role DmChk2 in the regulation of γ-irradiation-induced apoptosis in the developing eye in Drosophila.
Grp Does Not Influence the Dmp53-Induced Eye Phenotype.
Grp is the Drosophila homolog of Chk1. To investigate the interaction between Dmp53 and Grp in vivo, we generated transgenic flies overexpressing wild-type and DN-Grp. We generated a DN-Grp by mutating the conserved aspartic acid at position 130 to Ala (22). Overexpression of Grp in the posterior part of the eye imaginal disc resulted in a slight roughness and mild decrease in eye size (Fig. 4A) whereas DN Grp overexpression did not cause any detectable phenotype (Fig. 4B). Coexpression of either wild-type or DN-Grp with Dmp53 did not affect the Dmp53-induced phenotype (Fig. 4 C–E). Also, no change in the amount of acridine-orange-positive cells was observed (Fig. 4 F and G).
Fig 4.
Grp does not affect Dmp53 activity. (A–E) Scanning electron micrographs of eyes from flies overexpressing Grp (A), DN-Grp (B), Dmp53 (C), Grp and Dmp53 (D), or DN-Grp and Dmp53 (E) directed by GMR-Gal4. (Magnification: ×220.) (F–G) Acridine orange staining of eye discs overexpressing Grp and Dmp53 (F) or DN-Grp and Dmp53 (G). (Magnification: ×400.) (H) Expression of Grp and DN-Grp. Protein extracts from w118 (lane 1), Grp (lane 2), and DN-Grp (lane 3) flies crossed to armGal4 flies were analyzed by Western blot using anti-Chk1 Ab. Equal loading was confirmed by actin (Lower). (I) DN-Grp inhibits Grp kinase activity. Myc/His-tagged Grp, alone (lane 2) or together with increasing amounts of DN-Grp (lanes 4–6), immunoprecipitated from 293T cells, was incubated with synthetic Chk1/Chk2 substrate peptide, and the resulting phosphorylation was assessed by autoradiography. As a control (lane 7), the anti-Myc/His Ab was omitted in the immunoprecipitation step.
Eye imaginal discs from third-instar larvae overexpressing wild-type or DN-Grp were also irradiated, but no change in the number of apoptotic cells was noted compared to eye discs from wild-type larvae (data not shown). In addition, the Dmp53-associated eye phenotype was not altered when Dmp53 transgenic flies were crossed to a P element line [P(PZ)grp06034], in which the grp gene was heterozygously disrupted.
As neither the overexpression system with transgenic flies nor the loss-of-function system with mutant flies lacking one copy of the grp gene showed any alteration of the Dmp53 eye phenotype, we feel that Grp is likely not involved in the regulation of Dmp53.
Grp and DN-Grp transgenic flies were crossed to an arm-Gal4 transgenic line. Protein extracts from progeny larvae and control larvae were analyzed for Grp expression by Western blotting. Fig. 4H shows that both wild-type and DN-Grp proteins were expressed in vivo in transgenic flies.
Using an in-vitro kinase assay, we showed that the DN-Grp inhibited the function of its wild-type counterpart (Fig. 4I), demonstrating that the D130A mutation in the Grp protein functions as a true DN mutation inhibiting the kinase activity of the wild-type protein.
DmChk2 Controls Transcriptional Activity of p53.
The ability of Dmp53 to activate transcription in Drosophila S2 cells in the presence of wild-type or DN-DmChk2 and Grp was analyzed by using a human p53 responsive CAT-reporter construct (PG13-CAT) (18). Transfection of Dmp53 resulted in a dose-dependent increase in PG13-CAT reporter activity (Fig. 5A Upper). Cotransfection of DmChk2 with Dmp53 caused a further increase in reporter activity (Fig. 5A Upper). In contrast, expression of DN-DmChk2 interfered with Dmp53-mediated transcription (Fig. 5A Upper). In agreement with our genetic studies, wild-type and DN Grp constructs had no influence on Dmp53 transcriptional activity (Fig. 5B Upper). Dmp53 protein levels were unchanged by cotransfection with either wild-type or DN-DmChk2 or Grp constructs, indicating that the observed effects are not caused by changes in Dmp53 protein levels (Fig. 5 A and B Lower). These results establish DmChk2, and not Grp, as a regulator of Dmp53 transcriptional activity in Drosophila.
Ser-4 Is Important for Chk2 Regulation of Dmp53 Transcriptional Activity.
Phosphorylation and acetylation have been implicated in regulating mammalian p53 stability and transcriptional activity after DNA damage (27). Sites of particular interest are the Ser–Glu amino acid pairs at positions 15 and 37 of human p53, which can be phosphorylated on the Ser residues by members of the ATM family of DNA damage-responsive kinases like ATM, Chk1, or Chk2 (8, 12). Based on homology, the nearby Ser-4–Glu-5 pair in Dmp53 might be a target for one of these kinases.
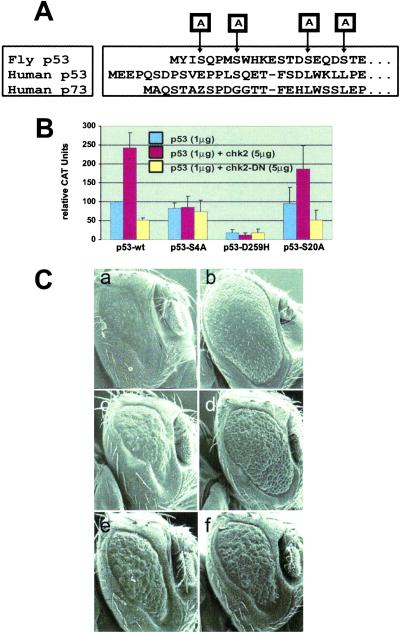
To define whether Dmp53 phosphorylation is part of the mechanism in the above-described regulation of Dmp53 by DmChk2, several point mutations (Dmp53-S4A, -S8A, -S16A, and -S20A) were introduced into the amino-terminal part of Dmp53 (Fig. 6A). Transfection of the various mutant Dmp53 molecules resulted in a similar increase in PG13-CAT-reporter activity as observed with the transfection of wild-type Dmp53 (Fig. 6B, p53-wt and data not shown). Cotransfection of DmChk2 with Dmp53-S20A caused a strong increase in reporter activity similar to wild-type Dmp53, whereas expression of DN-DmChk2 interfered with Dmp53-S20A (Fig. 6B, p53-S20A). Mutation of Ser-8 and Ser-16 also did not interfere with the transcriptional activation of Dmp53 by DmChk2 cotransfection (data not shown). These results suggest that Ser-8, -16, and -20 are not required for the regulation of Dmp53 activity by DmChk2 in Drosophila.
Fig 6.
Regulation of Dmp53 by DmChk2 via Ser-4. (A) Amino acid sequence alignment of Drosophila and hp53 and hp73. Arrows indicate mutated Ser residues. (B) Drosophila S2 cells were transfected with 1 μg of wild-type Dmp53, Dmp53-S4A, Dmp53-D259H, and Dmp53-S20A, alone (blue bars), and with 5 μg of wild-type or DN-DmChk2 (yellow bars) and p53 responsive PG13-CAT. (C) Scanning electron micrographs of eyes of a fly expressing Dmp53 (a and b), Dmp53-S20A (c and d), Dmp53-S4A (e and f) together with one copy of the DmChk2 (a, c, and e), or DN DmChk2 (b, d, and f) directed by GMR-Gal4. (Magnifications: ×220.)
Interestingly, when Dmp53-S4A was cotransfected with wild-type or DN DmChk2, no increase or decrease of the PG13-CAT-reporter activity was observed, indicating that Ser-4 might be crucial in the regulation of Dmp53 by DmChk2 (Fig. 6B, p53-S4A). The D259H point mutation did not induce any reporter activity and was not affected by cotransfection with either wild-type or DN DmChk2 (Fig. 6B).
The analysis was extended into the Drosophila eye. Transgenic flies expressing some of the Ser mutants of Dmp53 were constructed. Overexpression of the Dmp53 mutants S4A and S20A in the fly eye resulted in a small eye phenotype identical to the overexpression of wild-type Dmp53 (data not shown). Genetic epistasis was performed crossing these flies to transgenic flies overexpressing either wild-type or DN-DmChk2. In agreement with the results from S2 cells, Dmp53-S20A (Fig. 6 Cc and Cd) behaved indistinguishably from the wild-type Dmp53 (Fig. 6 Ca and Cb), whereas the S4A mutant resulted in a phenotype that was unaltered by coexpression with wild-type or DN-DmChk2 (Fig. 6 Ce and Cf). These results confirm that Ser-4 of Dmp53 is important for its regulation by DmChk2.
Discussion
Upon DNA damage, cells respond with cell cycle arrest and activation of genes that coordinate DNA repair. If these mechanisms fail, genomic instability and predisposition for the development of cancer is the consequence.
Central to the execution of the DNA damage response is p53. Regulation of p53 is coordinated mainly by two mechanisms: regulation of its stability and its activity. Ionizing irradiation induces phosphorylation of several amino-terminal amino acids of mammalian p53, a process that is mediated by both Chk1 and Chk2 (7, 8). Phosphorylation of human p53 at Ser-20 interferes with binding of murine double minute 2 protein (MDM2) to p53, thereby inhibiting degradation of p53 and leading to an increase in p53 protein levels (5, 8, 12). The mechanism underlying the regulation of p53 transcriptional activity is less clear, but phosphorylation of p53 could be involved in this process as well. In mammals, both regulatory mechanisms are tightly linked together and are difficult to differentiate. In Drosophila, genomewide searches have not identified an MDM2 homolog, allowing the investigation of the mechanism of p53-regulation independent of MDM2.
In this study, we demonstrate that DmChk2 is a potent activator of Dmp53. Expression of both molecules in the Drosophila eye led to a massive reduction in eye size caused by an increase in the number of apoptotic cells. Interestingly, the requirement for DmChk2 to activate Dmp53 was observed only upon overexpression or Dmp53 or after radiation treatment, as overexpression of DmChk2 or DN-DmChk2 alone did not show a phenotype. Du and coworkers (28) also have shown that flies homozygously deleted for DmChk2 are viable and lack obvious phenotype in the absence of irradiation. These observations suggest that endogenous Dmp53 may function primarily in a DNA damage response. A kinase-dead, DN-DmChk2 was able to almost fully rescue the p53-induced phenotype by inhibiting apoptosis in the eye imaginal discs. A functional interaction between Grp and Dmp53 could not be detected, suggesting that DmChk2 is the principal activator of Dmp53 in Drosophila. In an attempt to define the molecular mechanism of the observed interaction between Dmp53 and DmChk2, several point mutations were introduced into the amino-terminal part of the Dmp53 molecule and tested for their responsiveness to wild-type or DN-DmChk2. The transcriptional activity and the eye phenotype caused by Dmp53-S4A mutant were unaffected by coexpression with either wild-type or DN-DmChk2. These results indicate that Ser-4 of Dmp53 confers responsiveness to DmChk2.
We hypothesized that the regulation of Dmp53 by DmChk2 could be mediated by direct phosphorylation. However, in vitro DmChk2 kinase assays using wild-type or mutant glutathione S-transferase-p53 proteins as substrates did not detect any differences in phosphorylation between the various p53 molecules (data not shown). One explanation for this phenomenon is that DmChk2 does not directly phosphorylate Ser-4 and that a DmChk2-regulated kinase is involved in this process. Alternatively, DmChk2 may phosphorylate Dmp53 at multiple sites, preventing discrimination between wild-type and Dmp53-S4A phosphorylation levels.
Dmp53-S4A overexpressing flies exhibited a similar phenotype to Dmp53 wild-type transgenic flies. This finding is surprising given that DN-DmChk2 almost completely rescued the Dmp53-induced small eye phenotype. There may be several reasons for this. Other signals not related to DmChk2 that may or may not involve phosphorylation could be constitutively active, resulting in p53 stabilization and transcriptional activity. In this case, mutation of S4 could inhibit further activation of Dmp53 but would not prevent the constitutive activating signals present. Alternatively, introduction of the S4 mutation could introduce steric changes that confer increased stability to the p53 molecule. The increased stability could also account for the observed phenotype. A further possibility is that DmChk2 additionally activates a kinase that phosphorylates Dmp53. In this situation, mutation of Dmp53 Ser-4 would prevent direct activation of p53 but would not interfere with indirect activation. This also would explain why DN-DmChk2 could prevent Dmp53-mediated apoptosis.
We have shown that radiation-induced cell death is inhibited by DN-Dmp53 and DN-DmChk2 and that apoptosis resulting from Dmp53 overexpression can be inhibited by a kinase-dead form of DmChk2. DmChk2 regulation of Dmp53 activity is crucial for γ-radiation-induced apoptosis, consistent with data showing that DmChk2 functions upstream of Dmp53 (8). In favor for this view, Du and coworkers recently described DmChk2 null flies that are refractory to apoptosis upon irradiation, suggesting that DmChk2 controls Dmp53-induced apoptosis (28). However, we cannot rule out a scenario in which Dmp53 and DmChk2 make independent contributions to radiation-induced cell death, functioning in separate pathways.
As Drosophila most likely does not have an MDM2 gene, the MDM2-mediated p53 degradation pathway could have emerged at a later point in evolution. The data presented here suggest that MDM2-independent regulation of Dmp53 is mediated by DmChk2 and implies that DmChk1 does not function in this process. If MDM2-independent regulation of p53 is more ancient in evolutionary terms, then Chk2 may be the ancestral regulator of p53 activation. Thus, MDM2 and Chk1 probably emerged as regulators of p53 at a later evolutionary time.
The ability of DmChk2 to regulate transcriptional activity of Dmp53 suggests that in mammals Chk2 might be involved in control of both p53 stability and activity. Generation of mouse “knock-in” mutants for individual amino-terminal p53 phosphorylation sites should aid in resolution of apparent multiple roles of Chk2 in p53 regulation.
Acknowledgments
We thank Drs. Gerald M. Rubin and Michael H. Brodsky for generously providing the DN Dmp53 flies as well as the reagents for the CAT assay. We thank Dr. James Woodgett for the gift of Drosophila S2 cells. C.D.L. is supported by an Medical Research Council of Canada postdoctoral fellowship. V.S. is a recipient of a postdoctoral fellowship from the Cancer Research Institute, New York. M.P. is supported by a Feodor-Lynen Fellowship of the Alexander von Humboldt-Foundation.
Abbreviations
Dmp53, D. melanogaster p53
DmChk2, D. melanogaster Chk2
Grp, Grapes
ATM, ataxia telangiectasia mutated
CAT, chloramphenicol acetyltransferase
DN, dominant-negative
GMR-Gal4, glass multimer reporter
MDM2, murine double minute 2
References
- 1.Levine A. J. (1997) Cell 88, 323-331. [DOI] [PubMed] [Google Scholar]
- 2.Vousden K. H. (2000) Cell 103, 691-694. [DOI] [PubMed] [Google Scholar]
- 3.Walworth N. C. (2000) Curr. Opin. Cell. Biol. 12, 697-704. [DOI] [PubMed] [Google Scholar]
- 4.Zhou B. S. & Elledge, S. J. (2000) Nature (London) 408, 433-439. [DOI] [PubMed] [Google Scholar]
- 5.Chehab N. H., Malikzay, A., Appel, M. & Halazonetis, T. D. (2000) Genes Dev. 14, 278-288. [PMC free article] [PubMed] [Google Scholar]
- 6.Matsuoka S., Huang, M. & Elledge, S. J. (1998) Science 282, 1893-1897. [DOI] [PubMed] [Google Scholar]
- 7.Blasina A., de Weyer, I. V., Laus, W. H., Luyten, W. H., Parker, A. E. & McGowan, C. H. (1999) Curr. Biol. 9, 1-10. [DOI] [PubMed] [Google Scholar]
- 8.Hirao A., Kong, Y.-Y., Matsuoka, S., Wakeham, A., Ruland, J., Yoshida, H., Liu, D., Elledge, S. J. & Mak, T. W. (2000) Science 287, 1824-1827. [DOI] [PubMed] [Google Scholar]
- 9.Rhind N. & Russell, P. (2000) J. Cell Sci. 113, 3889-3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rhind N. & Russell, P. (1998) Curr. Opin. Cell. Biol. 10, 749-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Russell P. (1998) Trends Biochem. Sci. 23, 399-402. [DOI] [PubMed] [Google Scholar]
- 12.Shieh S.-Y., Ahn, J., Tamai, K., Taya, Y. & Prives, C. (2000) Genes Dev. 14, 289-300. [PMC free article] [PubMed] [Google Scholar]
- 13.Hari K. L., Santerre, A., Sekelsky, J. J., Makim, K. S., Boyd, J. B. & Hawley, R. S. (1995) Cell 82, 815-821. [DOI] [PubMed] [Google Scholar]
- 14.Fogarty P., Campbell, S. D., Aby-Shumays, R., Phalle, B. S., Yu, K. R., Uy, G. L., Goldberg, M. L. & Sullivan, W. (1997) Curr. Biol. 7, 418-426. [DOI] [PubMed] [Google Scholar]
- 15.Oishi I., Sugiyama, S., Otani, H., Yamamura, H., Nishida, Y. & Minami, Y. (1998) Mech. Dev. 71, 49-63. [DOI] [PubMed] [Google Scholar]
- 16.Higashitani A., Aoki, H., Mori, A., Sasagawa, Y., Takanami, T. & Takahashi, H. (2000) FEBS Lett. 485, 35-39. [DOI] [PubMed] [Google Scholar]
- 17.MacQueen A. J. & Villeneuve, A. M. (2001) Genes Dev. 15, 1674-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brodsky M. H., Nordstrom, W., Tsang, G., Kwan, E., Rubin, G. M. & Abrams, J. M. (2000) Cell 101, 103-113. [DOI] [PubMed] [Google Scholar]
- 19.Ollmann M., Young, L. M., Di Como, C. J., Karim, F., Belvin, M., Robertson, S., Whittaker, K., Demsky, M., Fisher, W. W., Buchman, A., et al. (2000) Cell 101, 91-101. [DOI] [PubMed] [Google Scholar]
- 20.Jin S., Martinek, S., Joo, W. S., Wortman, J., Mirkovic, N., Sali, A., Yandell, M. D., Pavletich, N. P., Young, M. W. & Levine, A. J. (2000) Proc. Natl. Acad. Sci. USA 97, 7301-7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohler G. & Milstein, C. (1975) Nature (London) 256, 495-497. [DOI] [PubMed] [Google Scholar]
- 22.Sanchez Y., Wong, C., Thoma, R. S., Richman, R., Wu, Z., Piwnica-Worms, H. & Elledge, S. J. (1997) Science 277, 1497-1501. [DOI] [PubMed] [Google Scholar]
- 23.Furnari B., Rhind, N. & Russell, P. (1997) Science 277, 1495-1497. [DOI] [PubMed] [Google Scholar]
- 24.Neufeld T. P., de la Cruz, A. F., Johnston, L. A. & Edgar, B. A. (1998) Cell 93, 1183-1193. [DOI] [PubMed] [Google Scholar]
- 25.Brand A. & Perrimon, N. (1993) Development (Cambridge, U.K.) 118, 401-415. [DOI] [PubMed] [Google Scholar]
- 26.Cho Y., Gorina, S., Jeffrey, P. D. & Pavletich, N. P. (1994) Science 265, 346-355. [DOI] [PubMed] [Google Scholar]
- 27.Lakin N. D. & Jackson, S. P. (1999) Oncogene 18, 7644-7655. [DOI] [PubMed] [Google Scholar]
- 28.Xu J., Xin, S. & Du, W. (2001) FEBS Lett. 508, 394-398. [DOI] [PubMed] [Google Scholar]