Abstract
A mouse model has been established to investigate the genetic determinism of host susceptibility to West Nile (WN) virus, a member of the genus flavivirus and family Flaviviridae. Whereas WN virus causes encephalitis and death in most laboratory inbred mouse strains after peripheral inoculation, most strains derived from recently trapped wild mice are completely resistant. The phenotype of resistance/susceptibility is determined by a major locus, Wnv, mapping to chromosome 5 within the 0.4-cM-wide interval defined by markers D5Mit408 and D5Mit242. We constructed a high resolution composite/consensus map of the interval by merging the data from the mouse T31 Radiation Hybrid map and those from the homologous region of human chromosome 12q, and found the cluster of genes encoding 2′-5′-oligoadenylate synthetases (2′-5′-OAS) to be the most prominent candidate. This cluster encodes a multimember family of IFN-inducible proteins that is known to play an important role in the established endogenous antiviral pathway. Comparing the cDNA sequences of 2′-5′-OAS L1, L2, and L3 isoforms, between susceptible and resistant strains, we identified a STOP codon in exon 4 of the gene encoding the L1 isoform in susceptible strains that can lead to a truncated form with amputation of one domain, whereas all resistant mice tested so far have a normal copy of this gene. The observation that WN virus sensitivity of susceptible mice was completely correlated with the occurrence of a point mutation in 2′-5′-OAS L1 suggests that this isoform may play a critical role in WN pathogenesis.
Flavivirus is a genus of the Flaviviridae family that comprises over 70 positive-sense, single-stranded, and enveloped RNA viruses, most of which are arthropod-borne (1). Mosquito-borne flaviviruses such as dengue (DEN), Japanese encephalitis (JE), yellow fever (YF), and West Nile (WN) viruses can cause epidemic outbreaks in humans, and patients infected may exhibit a wide range of acute diseases, from nonspecific febrile illness to severe hemorrhagic manifestations (DEN and YF) or encephalitic syndromes (JE and WN), sometimes associated with a high (up to 50%) fatality rate. The reasons why flaviviruses cause clinical manifestations only in a small percentage of infected individuals have not yet been clearly elucidated but are supposed to involve host-dependent genetic factors.
Over the past 5 years, WN fever has been an emerging concern for public health in Europe, in the Middle East, and more recently in the United States. In Israel and the United States, where the Isr98/NY99 variant of WN virus was frequently isolated during recent outbreaks, approximately 20% of infected persons developed febrile illness, with a high rate of mortality among patients with neurological symptoms (2, 3). This result suggests that susceptibility and sensitivity to WN virus infections might depend, at least in part, on host genetic factors (4). In the present study we investigated the influence of the host genetic constitution on the severity of WN virus infection, by using the viral strain IS-98-ST1, a variant closely related to strain Isr98/NY99, which has the capacity to kill adult mice of most laboratory strains after peripheral inoculation. Our data show that, within the genus Mus, susceptibility to WN experimental infections is correlated with the occurrence of a point mutation resulting in the truncation of the 2′-5′-oligoadenylate synthetase (2′-5′-OAS) L1 isoform, which suggests that this enzyme may be critical in WN pathogenesis through an effect restricting viral replication in target tissues.
Materials and Methods
WN Virus Production and Titration.
WN virus strain IS-98-ST1 (or Stork/98) was isolated from a stork in Israel, in 1998, by C. Bannet and M. Malkinson (Kimron Veterinary Institute, Israel; refs. 5 and 6). This strain displays less than 0.3% sequence divergence from the variant NY99 of WN virus (GenBank accession no. AF481864; ref. 6). Production of WN virus on mosquito Aedes pseudoscutellaris cell monolayers, purification, and virus titration by focus immunodetection assay (FIA) were performed as previously described (7). Infectivity titers were expressed as focus forming units (FFU). To quantify the amount of infectious virus produced in mouse brain, tissue samples were prepared as 10% suspensions, and virus titers were expressed as FFU/g of brain tissue (8).
Mouse Strains and Crosses.
Mice of inbred strains BALB/cByJ@Ico, DBA/2J@Ico, and C57BL/6J@Ico were purchased from Charles River France Laboratories. Mice of inbred strain PL/J were purchased from The Jackson Laboratory. Mice of inbred strains NZB/Ola and NZW/Ola were from Olac (Bichester, U.K.). Mice of strains 129/Sv/Pas, C3H/He/Pas, and DDK/Pas were bred in our facilities as were mice of strains WMP/Pas, PWK/Pas, MBT/Pas, MAI/Pas, SEG/Pas, and STF/Pas, which all were derived from wild specimens of different species or subspecies of the Mus genus. All these mice are highly inbred strains (F ≥50). The BALB/c-MBT (N7) congenic strain was produced by the marker-assisted selective introgression of a short segment of MBT/Pas Chromosome (Chr) 5 flanked by marker D5Mit242 and D5Mit158, into a BALB/c background. For the analysis of the genetic determinism of susceptibility (S) or resistance (R) to WN virus experimental infections, four backcross progenies were bred by crossing (BALB/c × MAI), (BALB/c × MBT), (C57BL/6 × MBT), or (C57BL/6 × MAI) F1 males to BALB/c or C57BL/6 laboratory females, respectively.
Virulence Determination in Mice.
Groups of 6-wk-old mice were inoculated either i.p. with 100 μl, or intracerebrally with 20 μl of serial dilutions of WN virus in PBS with 0.2% BSA. Mice were observed daily for 21 days, and deaths were recorded. Fifty percent lethal dose (LD50) values were calculated by using the method of Reed and Muench (9).
Immunodetection of WN Viral Antigen in the Central Nervous System (CNS).
Brains were dissected intact and rapidly frozen on dry ice before sectioning on a cryostat (Jung Frigocut, Nussloch, Germany), according to the parasagittal axis. Tissue sections (16-μm thick) were then air-dried and fixed for 30 min in freshly prepared, phosphate-buffered 3.7% formaldehyde before being processed for immunofluorescence (8). Serum specimens from resistant mice i.p. inoculated with IS-98-ST1 virus were used as the primary antibody after specificity controls on infected/uninfected brain sections. FITC-conjugated horse anti-mouse antibody (Vector Laboratories) was used to detect specifically bound primary antibodies. Sections were then mounted in Vectashield Medium (Vector Laboratories) and observed under a fluorescence microscope (Leica DMRB, Bensheim, Germany).
Identification and Mapping of the Genetic Determinants for Susceptibility/Resistance After Experimental Infection with WN Virus.
DNA samples were prepared from small pieces of tail taken from each mouse of the four backcross progenies before experimental infections. Then, these DNA samples were typed with a series of 81 microsatellite markers evenly distributed over the whole genome (genome scan). Finally, once challenged with WN virus, the genotypes were matched with the R/S phenotypic pattern, and the results were analyzed with the gene link software (10).
Sequence Analysis of L1 Isoform of 2′-5′-OAS.
Primary cultures of neurons derived from the brains of 14-day mouse embryos were used to prepare cDNA. The cells were induced by IFN-α at a dose of 20 units/ml (GIBCO/BRL). The cDNA fragment of the L1 isoform of 2′-5′-OAS was PCR amplified by using the following primers: 5′-AGGTAAAAGCTGGACCTAGG-3′ and 5′-TGTTGGTGCAGGTATTCAGAGAC-3′. Genomic fragments of exon 4 of the gene encoding the 2′-5′-OAS L1 isoform were PCR amplified by using 5′-ACACAGTGTCCATCTCAACCA-3′ and 5′-TGTTGGTGCAGGTATTCAGAGAC-3′. Amplification products were purified on gels and cloned into a dT-tailed pCR4-TOPO vector (Invitrogen). At least, three independent clones were sequenced for each fragment. The cDNA sequence of the 2′-5′-OAS L1 isoform was deposited in GenBank (accession nos: BALB/c, AF466822; MBT/Pas, AF466823).
Results
In this study, we investigated the influence of host genetic determinants on WN virus pathogenicity in the mouse species. To address this question, we developed a strategy in which adult mice were i.p. inoculated with the highly virulent strain IS-98-ST1, which has an i.p. LD50 of 10 FFU in 6-wk-old outbred Swiss mice (5). This strain has the same level of peripheral virulence as NY99 for adult mice (5, 11). As data in Table 1 show, i.p. inoculation of 1,000 FFU (100 i.p. LD50) of the WN virus strain IS-98-ST1 caused death in all 6-wk-old inbred BALB/c mice. Although a WN-specific antibody response was observed by day 7 of infection, infected BALB/c mice did not survive for more than 13 days (mean day of death, 9.5 ± 1.5 days). The spread of WN virus in brain tissues was assessed by virus titration (Table 1). Infectious IS-98-ST1 was detected in brain tissues after 5 days of infection, and the amounts of virus peaked at 109 FFU/g by day 7. When brain tissue sections were assayed for the presence of viral antigens by immunostaining, WN virus replication was detected mostly in the spinal cord, thalamus, cerebellum, and hippocampus (data not shown). These data are consistent with the notion that WN wild-type strain IS-98-ST1 is neuroinvasive and neurovirulent for adult mice (5). Further experiments showed that i.p. inoculation of 1,000 FFU of IS-98-ST1 caused 100% mortality in all classical laboratory inbred strains tested except PL/J, whereas no morbidity or mortality was observed in 6-wk-old mice from six totally unrelated inbred strains derived from wild ancestors of the Mus m. domesticus (WMP/Pas), Mus m. musculus (MAI/Pas, MBT/Pas, PWK/Pas), or Mus spretus (SEG/Pas, STF/Pas) species challenged under the same conditions (Table 2). The high levels of anti-WN antibody produced in surviving animals indicated that WN virus did replicate in resistant mouse strains.
Table 1.
WN virus strain IS-98-ST1 is neuroinvasive and neurovirulent for BALB/c mice
| Day of infection | Mortality, no. of dead/ no. of infected mice | Viral titer in brain, log FFU/g | WN-specific antibody, O.D. 450 nm |
|---|---|---|---|
| 1 | 0/12 | <2 | 0.20 ± 0.05 |
| 3 | 0/12 | <2 | 0.20 ± 0.05 |
| 5 | 0/12 | 3.5–6.8 | 0.30 ± 0.10 |
| 7 | 0/12 | 9.3 | 0.60 ± 0.10 |
| 9 | 9/12 | 2–4 | 0.65 ± 0.01 |
| 12 | 11/12 | NA | NA |
| 13 | 12/12 | NA | NA |
Six-week-old BALB/c mice in a group of 12 were i.p. inoculated with 1,000 FFU of WN virus strain IS-98-ST1. Mice were observed daily, and mortality was recorded.
Brain tissues were extracted from three WN virus-infected mice and weighted. Brain tissues were prepared as 10% (wt/vol) suspensions, and their infectivity was titrated by FIA (8). Detection threshold, 100 FFU/g.
Three WN virus-infected mice were bled to evaluate the WN-specific antibody response. Antibody titer was determined by immunoglobulin capture enzyme-linked immunoabsorbent assay (ELISA). Briefly, microtitration plaques were coated with 100 μl of highly-purified WN virus strain IS-98-ST1 (106 FFU) in PBS. After saturation, mouse sera diluted at 1:100 were incubated with WN virus. Peroxidase-conjugated goat anti-mouse IgG (H+L) antiserum (BIOSYS) was used as the second antibody, and ELISA was performed with TMB microwell peroxidase substrate system (Kirkegaard & Perry Laboratories). Optical density (OD ± SD) was measured at 450 nm.
NA, Not applicable.
Table 2.
Inheritance of resistance to WN virus
| Mouse strains | Mortality, no. of dead/ no. of infected mice | Days of death (extremes) |
|---|---|---|
| BALB/c | 6/6 | 8–10 |
| C57BL/6 | 6/6 | 7–10 |
| MAI/Pas | 0/6 | NA |
| MBT/Pas | 0/6 | NA |
| (C57BL/6 × MAI)F1 | 0/6 | NA |
| (C57BL/6 × MBT)F1 | 0/6 | NA |
| (BALB/c × MAI)F1 | 0/6 | NA |
| (BALB/c × MBT)F1 | 0/6 | NA |
| (C57BL/6 × MAI)F1 × C57BL/6 | 16/42 | 7–13 |
| (C57BL/6 × MBT)F1 × C57BL/6 | 8/25 | 10–12 |
| (BALB/c × MAI)F1 × BALB/c | 19/55 | 7–13 |
| (BALB/c × MAI)F1 × BALB/c | 31/81 | 7–12 |
Four sets of F1 hybrids were produced by crossing males of resistant strain MAI or MBT to females of susceptible strain BALB/c or C57BL/6. Four sets of backcross mice were generated by crossing the F1 males to females of the susceptible parental strain.
Six-week-old BALB/c mice were i.p. inoculated with 1,000 FFU of WN virus strain IS-98-ST1. Survival was recorded for 21 days.
NA, Not applicable.
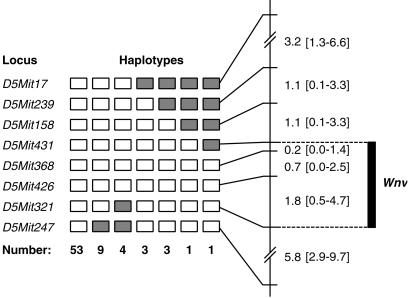
Based on these observations, we undertook experiments to unravel the genetic determinism of the resistance/susceptibility (R/S) phenotype of WN virus infection. To achieve this goal, a total of 203 progeny from four backcrosses involving strains BALB/c or C57BL/6 (susceptible) and MBT or MAI (resistant) were challenged by the same i.p. route, with the same 1,000-FFU dose of IS-98-ST1 WN virus, and mice were observed every day for 21 days for signs of illness or death. Matching genotypes with R/S phenotypes allowed us to identify and map a major genetic determinant on Chr 5, for which the symbol Wnv (West Nile virus) was coined. Wnv maps to the interval flanked by markers D5Mit431 and D5Mit321 (Fig. 1), a region where a locus determining the phenotype of resistance or susceptibility to flavivirus (Flv) had previously been mapped (12–14).
Fig 1.
Analysis of 74 haplotypes from backcross mice that died after experimental infection with WN virus allowed localization of the genetic determinant for resistance (symbol Wnv) in a 2.7-cM-long segment of mouse chromosome 5. Open rectangles symbolize homozygosity for the microsatellite marker in question, filled rectangles symbolize heterozygosity. Genetic distances between molecular markers are given with 95% confidence intervals. They were calculated by using the whole backcross progeny (203 mice).
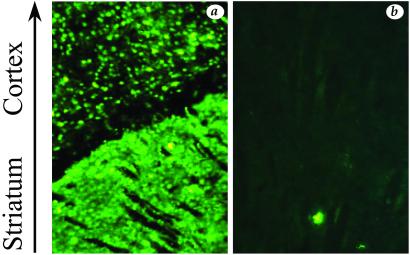
Flv controls susceptibility to neurotropic flaviviruses in mice, and it is known that the protection conferred by Flvr, the resistant allele at the Flv locus, correlates with the restriction of virus replication in the CNS (12–15). We were therefore interested in determining whether the WN virus replicates differently within the CNS of susceptible and resistant strains. For this experiment, we chose to challenge adult BALB/c mice and adult mice of the BALB/c-MBT resistant congenic strain. After intracranial inoculation with 1,000 FFU of IS-98-ST1, all BALB/c mice died from encephalitis within 8 days of infection, whereas all BALB/c-MBT congenic mice survived despite transient neurological symptoms. Immunohistological analysis of infected BALB/c brains revealed that WN virus-infected neural cells were largely distributed in the cortex and striatum of BALB/c mice (Fig. 2a), whereas only a small number of neural cells were positive for WN antigens in the CNS of BALB/c-MBT mice (Fig. 2b). This finding correlated with the low amounts of infectious virus in the brains of infected congenic mice (average titer, 5 × 106 FFU/g) compared with those of BALB/c mice (average titer, 5 × 109 FFU/g). We concluded that the survival of WN virus-infected mice may be correlated with the restriction of virus replication in target tissues.
Fig 2.
Distribution of viral antigen-positive cells within WN virus-infected brain. BALB/c (a) and BALB/c-MBT (N7) congenic (b) mice were i.c. inoculated with 1,000 FFU of IS-98-ST1. Day 5 of injection, mouse brains were prepared for frozen sectioning and assayed for the presence of WN antigens by indirect fluorescence-antibody assay. Tissue sections were immunostained with anti-WN polyclonal antibody. Magnification, ≈×40.
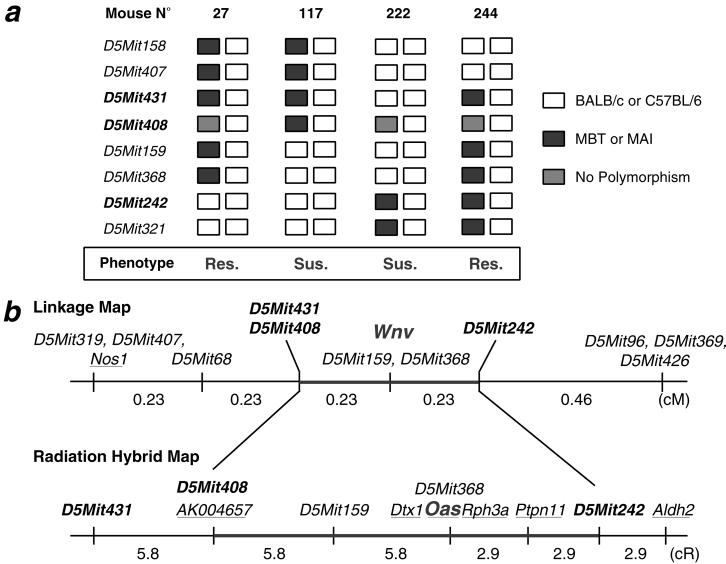
To reduce the genetic interval containing the Wnv locus and thereby facilitate the identification of candidate genes, mice recombinant in the 2.7 cM critical region were crossed again to the susceptible parent (second backcross or N3), and groups of 10 to 12 progeny from each of these informative recombinant mice were challenged by using the same i.p. inoculation protocol as mentioned above (Fig. 3a). This strategy allowed us to localize the genetic determinant for WN resistance within a reduced interval of 0.4 cM flanked by the microsatellite markers D5Mit408 and D5Mit242 (Fig. 3b).
Fig 3.
(a) Mice from (BALB/c × MBT)F1 × MBT or (C57BL/6 × MAI)F1 × MAI backcross progenies, with a Chr 5 recombinant in the interval where the Wnv locus maps (flanked by microsatellite markers D5Mit431 and D5Mit321), were identified and used for the generation of a second backcross generation (N3) by crossing with their susceptible parental strain (BALB/c or C57BL/6). Ten of twelve mice of this N3 generation were bred, then i.p. challenged with 1,000 FFU of IS-98-ST1 virus. Matching the haplotype constitutions to the phenotype of resistance/susceptibility (Res./Sus.) allowed reduction of the critical region to the interval flanked by markers D5Mit431 and D5Mit242 (0.4 cM). (b) Several genes mapping to the region flanked by markers D5Mit431 and D5Mit242, harboring the Wnv locus, were identified in the mouse genetic map established from the T31 Radiation Hybrid (RH) panel. Markers and Aldh2, which are mapped both in mouse and man, allowed in turn the establishment of an exhaustive list of the gene in the region (Table 3).
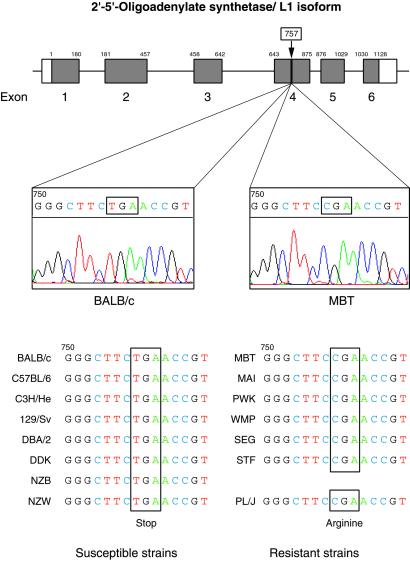
Combining the data from the mouse T31 Radiation Hybrid mapping resource (16) with the same flanking markers and merging information from the homologous region of human chromosome 12q (17) with our own data, we were able to establish a high-resolution consensus map of the critical region harboring Wnv. Among the 13 genes identified in this region (Table 3), the cluster encoding the family of 2′-5′-OAS appeared as a front line candidate because these genes, which are IFN-induced and double-stranded RNA-activated, encode efficient antiviral molecules (18–22). Analysis of the genomic sequences of the genes encoding the L1, L2, and L3 isoforms of 2′-5′-OAS (18, 19) of BALB/c and MBT/Pas mice revealed the presence of a STOP codon in exon 4 of the gene encoding the L1 form of 2′-5′-OAS in mice of the susceptible strain BALB/c, which was absent in the orthologous exon in the resistant strain MBT (Fig. 4). This STOP codon occurred in all (eight) susceptible strains tested in our challenge but was absent from all (six) resistant strains derived from wild specimens. The replacement of an Arg codon by an OPAL codon results in the truncation of the 2′-5′-OAS L1 isoform, with amputation of one of the two presumptive functional domains of the molecule. Pursuing our survey of R/S phenotypes among laboratory inbred strains, we also made the interesting observation that the PL/J strain, which was derived from the ancestral strain “Princeton Rockefeller” identified as resistant in former experiments by Sabin (23) and which also appeared to be the only laboratory strain tested by us and classified as resistant to WN virus in our assay, turned out to be the only one with a non-mutant exon 4 in the L1 encoding sequence (Fig. 4). Thus, there is an absolute correlation between the occurrence of a STOP codon in exon 4 of the gene encoding the L1 isoform of 2′-5′-OAS and the R/S phenotype in a set of 15 totally unrelated inbred strains. This finding suggests that this isoform plays a crucial role in the determinism of susceptibility to WN virus infection in mice.
Table 3.
Genes known to map within the human chromosomal segment (12q24) homologus to the mouse WNV interval on Chr 5
| Human locus | GenBank | Name | Locus link | OMIM | UniGene | Mouse locus | GenBank |
|---|---|---|---|---|---|---|---|
| FLN29 | AB007447 | FLN29 gene product | 10906 | — | Hs.5148 | — | — |
| RPL6 | X69391 | Ribosomal protein L6 | 6128 | 603703 | Hs.174131 | Rpl6 | AF374195 |
| PTPN11 | D13540 | Protein tyrosine phosphatase, non-receptor type 11 | 5781 | 176876 | Hs.22868 | Ptpn11 | D84372 |
| KIAA0985 | AB023202 | Rabphilin-3A | 22895 | — | Hs.21239 | Rph3a | D29965 |
| OAS1¶ | 2′-5′oligoadenylate synthetase 1 | 4938 | 164350 | Hs.82396 | Oas1 | ||
| OAS3 | 2′-5′oligoadenylate synthetase 3 | 4940 | 603351 | Hs.56009 | — | — | |
| OAS2 | 2′-5′oligoadenylate synthetase 2 | 4939 | 603350 | Hs.264981 | — | — | |
| DTX1 | AF053700 | Deltex (Drosophila) homolog 1 | 1840 | 602582 | Hs.124024 | Fxit1,Dtx1 | U38252 |
| RASAL1 | AF086713 | RAS protein activator like 1 | 8437 | 604118 | Hs.198312 | Rasal1 | AF086714 |
| KIA1169 | AK000619 | Two-pore channel 1, homolog | 53373 | — | Hs.26440 | — | — |
| SDS | J05037 | Serine dehydratase | 10993 | 182128 | Hs.76751 | Sds | AF328927 |
| LHX5 | AF291181 | LIM homeobox protein 5 | 64211 | 605992 | Hs.302029 | Lhx5 | U61155 |
| KIAA0682 | AL117547 | Hypothetical protein | 9904 | — | Hs.7482 | (RIKEN cDNA) | AK004657 |
¶The genes encoding 2′-5′-OAS appear to be strong candidates for Wnv.
Fig 4.
The C-T transition in the Arg codon at position 757 results in the occurrence of an opal (stop) codon in exon 4 of the gene encoding L1 isoform of 2′-5′-OAS in laboratory mice. This mutation results in the truncation of the L1 isoform, with amputation of one of its presumptive functional domains (GenBank accession nos: BALB/c, AF466822; MBT/Pas, AF466823). The structure of the gene encoding 2′-5′-OAS L1 isoform, with its six exons, appears at the top of Fig. 4. Scale for exons is 10 times larger than for introns. Susceptible strains, listed on the left side, are all classical laboratory inbred strains whereas most resistant strains, listed on the right side, are inbred strains derived from recently trapped wild specimens. Strain PL/J is the only exception known so far of a resistant classical laboratory inbred strain.
Discussion
To improve our understanding of the genetic determinism of susceptibility to flaviviruses, we have established a mouse model for WN virus infection in which IS-98-ST1, a strain closely related to the new variant Isr98/NY99, was injected i.p. into 6-wk-old animals. The advantage of our system model is that adult laboratory inbred mice were highly susceptible to a low-dose virus inoculum, a pattern that is different from the widely found notion that peripheral injection of wild-type strains of flaviviruses in adult mice often fails to result in morbidity and mortality over a wide dose range. So far, most of the experimental studies aimed at the elucidation of the genetic determinism of resistance/susceptibility to infections with flaviviruses were performed by inoculating mice i.p. with mouse-adapted strains of flaviviruses. However, it is important to note that mouse-neuroadapted strains of flaviviruses can significantly differ in their pathogenicity from the virus populations circulating in nature, and this finding may influence the susceptibility to infection by providing a selective advantage to highly neurovirulent variants.
Our genetic data demonstrate that the severity of WN virus experimental infection of mice is controlled by a major genetic determinant, Wnv. We were able to map the Wnv locus at high resolution, within an interval of 0.4 cM, in the vicinity of the region where the locus for flavivirus resistance (Flv) had previously been mapped (12–14). Thirteen genes map to this critical 0.4 cM interval of mouse Chr 5 harboring the Wnv locus (Table 3). Based on their known function, some of these genes were not considered strong candidates and were eliminated. Conversely, and considering their cellular functions, the cluster of genes encoding the 2′-5′-OAS were potential candidates and were analyzed in detail. After sequencing the transcribed regions encoding the L1, L2, and L3 isoforms, in both BALB/c and MBT mice, we found several single nucleotide polymorphisms (SNPs), but most of these differences appeared to be neutral mutations. Several other differences in the sequences were associated with amino acid substitutions in the encoded proteins but appeared to differ from one strain to the next irrespective of the phenotype of resistance or susceptibility. Finally, only two differences were perfectly correlated with the phenotype of resistance/susceptibility among the 15 unrelated mouse inbred strains studied: (i) the T-C transition at position 757, resulting in the above mentioned nonsense or STOP mutation and (ii) an A-G transition at position 797, i.e., downstream of the STOP codon. The most likely hypothesis to account for the presence of these two polymorphisms is that all of the classical laboratory strains we tested, except strain PL/J, inherited the same segment of Chr 5 from a common ancestor. Such a situation is not uncommon among mouse laboratory strains and was already observed by Staeheli and colleagues when they identified the genetic determinism of susceptibility to orthomyxoviruses infections (24, 25).
The perfect correlation between susceptibility and the occurrence of a STOP codon in exon 4 of the L1 isoform encoding gene supports the hypothesis that the truncated, and presumably inactive, form of 2′-5′-OAS L1 is the cause of the innate susceptibility to WN virus infection. This discovery, which is compatible with susceptibility behaving as a recessive trait, also indicates that, although there are several independent isoforms of 2′-5′-OAS, L1 appears to be a particularly important one. Investigating the genetic polymorphism of the human orthologous region might explain, at least in some cases, the differential susceptibility to infection and pathological manifestations in individuals exposed to flaviviruses in endemic regions.
Acknowledgments
We thank colleagues of Transverse Research Program 21 for their interest in this project and Drs. Philip Avner and Charles Babinet for their comments and suggestions. This work was funded by the Transverse Research Programs (Institut Pasteur) and Program de Recherche Fondamentale en Microbiologie et Maladies Infectieuses et Parasitaires, Ministère de l'Education Nationale, de la Recherche et de la Technologie, France. T.M. and M.L. are postdoctoral fellows at the Institut Pasteur.
Abbreviations
WN virus, West Nile virus
2′-5′-OAS, 2′-5′-oligoadenylate synthetase
CNS, central nervous system
Wnv, a locus determining resistance to West Nile virus
Flv, a locus determining resistance to flavivirus
FFU, focus forming unit
References
- 1.Monath T. P. & Heinz, F. X. (1996) in Virology, eds. Fields, B. N., Knipe, D. N., Howley, P. M., Chanock, R. M., Melnick, J. L., Monath, T. P., Roizman, B. & Strauss, S. E. (Lippincott-Raven, Philadelphia), pp. 961–1034.
- 2.Sampson B. A., Ambrosi, C., Charlo, A., Reiber, K., Veress, J. F. & Armbrustmacher, V. (2000) Hum. Pathol. 31, 527-531. [DOI] [PubMed] [Google Scholar]
- 3.Petersen L. R. & Roehrig, J. T. (2001) Emerging Infect. Dis. 7, 611-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sangster M. Y. & Shellam, G. R. (1986) Curr. Top. Microbiol. Immunol. 127, 313-318. [DOI] [PubMed] [Google Scholar]
- 5.Deubel V., Fiette, L., Gounon, P., Drouet, M.-T., Khun, H., Huerre, M., Banet, C., Malkinson, M. & Desprès, P. (2001) Ann. N. Y. Acad. Sci. 951, 195-201. [DOI] [PubMed] [Google Scholar]
- 6.Malkinson M., Banet, C., Weisman, Y., Pokamunski, S., King, R., Drouet, M.-T. & Deubel, V. (2002) Emerging Infect. Dis. 8, 392-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desprès P., Frenkiel, M.-P. & Deubel, V. (1993) Virology 196, 209-219. [DOI] [PubMed] [Google Scholar]
- 8.Desprès P., Frenkiel, M.-P., Ceccaldi, P.-E, Duarte dos Santos, C. N. & Deubel, V. (1998) J. Virol. 72, 823-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reed L. J. & Muench, H. (1938) Am. J. Hyg. 27, 493-497. [Google Scholar]
- 10.Montagutelli X. (1990) J. Hered. 81, 490-491. [DOI] [PubMed] [Google Scholar]
- 11.Pletnev G. A., Putnak, R., Speicher, J., Wagar, E. J. & Vaughn, D. W. (2002) Proc. Natl. Acad. Sci. USA 99, 3036-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sangster M. Y., Heliams, D. B., Mackenzie, J. S. & Shellam, G. R. (1993) J. Virol. 67, 340-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sangster M. Y., Urosevic, N., Mansfield, J. P., Mackenzie, J. S. & Shellam, G. R. (1994) J. Virol. 68, 448-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Urosevic N., Mansfield, J. P., Mackenzie, J. S. & Shellam, G. R. (1995) Mamm. Genome 6, 454-458. [DOI] [PubMed] [Google Scholar]
- 15.Urosevic N., van Maanen, M., Mansfield, J. P., Mackenzie, J. S. & Shellam, G. R. (1997) J. Gen. Virol. 78, 23-29. [DOI] [PubMed] [Google Scholar]
- 16.Rowe L. B., Barter, M. E. & Eppig, J. T. (2000) Genomics 69, 27-36. [DOI] [PubMed] [Google Scholar]
- 17.Lander E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2000) Nature (London) 409, 860-921. [Google Scholar]
- 18.Ichii Y., Fukunaga, R., Shiojiri, S. & Sokawa, Y. (1986) Nucleic Acids Res. 14, 10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rutherford M. N., Kumar, A., Nissim, A., Shebath, J. & Williams, B. R. (1991) Nucleic Acids Res. 19, 1917-1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Player M. R. & Torrence, P. F. (1998) Pharmacol. Ther. 78, 55-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castelli J., Wood, K. A. & Roule, R. J. (1998) Biomed. Pharmacother. 52, 386-390. [DOI] [PubMed] [Google Scholar]
- 22.Rebouillat D. & Hovanessian, A. G. (1999) J. Interferon Cytokine Res. 19, 295-308. [DOI] [PubMed] [Google Scholar]
- 23.Sabin A. B. (1952) Ann. N. Y. Acad. Sci. 54, 936-944. [DOI] [PubMed] [Google Scholar]
- 24.Staeheli P., Haller, O., Boll, W., Lindenmann, J. & Weissmann, C. (1986) Cell 44, 147-158. [DOI] [PubMed] [Google Scholar]
- 25.Staeheli P., Grob, R., Meier, E., Sutcliffe, J. G. & Haller, O. (1988) Mol. Cell. Biol. 8, 4518-4523. [DOI] [PMC free article] [PubMed] [Google Scholar]