Abstract
Amplification of chromosomal band 11q13 is a common event in human cancer. It has been reported in about 45% of head and neck carcinomas and in other cancers including esophageal, breast, liver, lung, and bladder cancer. To understand the mechanism of 11q13 amplification and to identify the potential oncogene(s) driving it, we have fine-mapped the structure of the amplicon in oral squamous cell carcinoma cell lines and localized the proximal and distal breakpoints. A 5-Mb physical map of the region has been prepared from which sequence is available. We quantified copy number of sequence-tagged site markers at 42–550 kb intervals along the length of the amplicon and defined the amplicon core and breakpoints by using TaqMan-based quantitative microsatellite analysis. The core of the amplicon maps to a 1.5-Mb region. The proximal breakpoint localizes to two intervals between sequence-tagged site markers, 550 kb and 160 kb in size, and the distal breakpoint maps to a 250 kb interval. The cyclin D1 gene maps to the amplicon core, as do two new expressed sequence tag clusters. We have analyzed one of these expressed sequence tag clusters and now report that it contains a previously uncharacterized gene, TAOS1 (tumor amplified and overexpressed sequence 1), which is both amplified and overexpressed in oral cancer cells. The data suggest that TAOS1 may be an amplification-dependent candidate oncogene with a role in the development and/or progression of human tumors, including oral squamous cell carcinomas. The approach described here should be useful for characterizing amplified genomic regions in a wide variety of tumors.
Increasing gene dosage through DNA amplification is a common feature of many tumors and results in up-regulation of tumor-promoting genes (1). Chromosomal band 11q13 seems to be one of the most frequently amplified regions in human cancer (2) and is associated with a poor prognosis (3). Amplification of this region has been reported in approximately 15% of breast carcinomas, 13% of lung cancers, 21% of bladder tumors, and 50% of esophageal cancers (4–7). We and others have observed that amplification of chromosomal band 11q13 occurs in the form of a homogeneously staining region in about 45% of oral squamous cell carcinomas (OSCC) and squamous cell carcinomas of the head and neck (SCCHN; refs. 7–9).
Substantial effort has been devoted to the physical mapping of band 11q13 by fluorescence in situ hybridization (FISH), long-range restriction mapping, and Southern blot analysis (10–15). Despite intensive effort, a comprehensive physical map of the 11q13 amplicon has not been published. More than 10 genes are known to reside in the 11q13 amplicon, including CCND1, FGF3, FGF4, and EMS1, but only CCND1 and EMS1 have been reported to be amplified and overexpressed consistently and, thus, thought to play a role in driving 11q13 amplification (5, 6).
To characterize further the 11q13 amplicon in OSCC, we used a technique called quantitative microsatellite analysis (QuMA; ref. 16). Here, we report the fine mapping of the 11q13 amplicon and the cDNA sequence and genomic structure of a previously uncharacterized gene, TAOS1 (tumor amplified and overexpressed sequence 1), and present evidence that it also may be important in driving amplification of 11q13.
Materials and Methods
Cell Culture.
Thirty OSCC cell lines developed from tumors removed from consenting patients who had not been treated previously were examined in this study (S.M.G., J. K. Reddy, S. Comsa, K. M. Rossie, C. M. Lese Martin, M. Shuster, B. N. Appel, R. Wagner, E. N. Myers, and J. T. Johnson, unpublished data, and Table 1, which is published as supporting information on the PNAS web site, www.pnas.org). The cells were cultured in MEM with Earle's salts supplemented with nonessential amino acids (Invitrogen), l-glutamine, gentamicin, and 10% (vol/vol) FBS (Irvine Scientific). Normal human oral keratinocytes (NHOK) were obtained from patients undergoing uvulopalatopharyngoplasty at the University of Pittsburgh Medical Center (consent obtained through the Oral Cancer Center at the University of Pittsburgh under Institutional Review Board Guidelines) and cultured as described (17).
Nucleic Acid Extraction and Analysis.
Cell line DNA was isolated by using the PureGene kit (Gentra Systems), and a secondary purification was carried out with the DNeasy tissue kit (Qiagen, Chatsworth, CA).
RNA was extracted by the Trizol reagent method according to the manufacturer's instructions (Invitrogen), purified by the RNeasy mini kit (Qiagen), and quantified by spectrophotometry. For Northern blot analysis, an aliquot of the purified RNA was used to isolate mRNA using the Oligotex mRNA mini kit (Qiagen). The mRNA samples (1 μg of each) were electrophoresed through a formaldehyde-agarose gel (0.9%), transferred to Hybond-N Nylon membranes (Amersham Pharmacia), and fixed to the membrane by UV crosslinking in a Stratalinker (Stratagene).
OSCC cell line Northern blots and multiple tissue Northern (MTN) blots (CLONTECH) were hybridized with 32P-labeled probes for the two new expressed sequence tags (EST). The EST clones, AI885296 (I.M.A.G.E. Consortium CloneID 2432346) and AA885110 (I.M.A.G.E. Consortium CloneID 1468477; ref. 18), were obtained from Research Genetics (Huntsville, AL) and were sequenced by us. Hybridization probes were made by PCR amplification (5′-CCAGCCCATAAATGAGTTC–3′, and 5′-GAGCATTTGTGCCTGAAG-3′ for AI885296; 5′-GCCAGAATTCGGTTCGAG-3′, and 5′-AACCCACAGGAACCTCAG-3′ for AA885110) of nonredundant sequence regions of the EST clones. After hybridization, the membrane was washed and exposed in a PhosphorImager (Molecular Dynamics).
Physical Map.
The bacteria artificial chromosome (BAC) contig was constructed by using data in the human high throughput genomic sequences (HTGS) division of GenBank, nonredundant nucleotide sequence Databases, and the Celera Human Genome Database (http://publication.celera.com/). Physical mapping data on the microsatellite loci [(CA)n] at 11q13 were obtained from Daniela S. Gerhard (Washington University, St. Louis, MO; www.genetics.wustl.edu/gerhard/SCW11/11qMarkers.html), the Human Genome Database (GDB, www.gdb.org), or directly from the human chromosome 11q13 genome sequence. BAC clones from the 11q13 amplicon were identified by screening the HTGS database and the Washington University human BAC fingerprint contig (FPC) database (http://genome.wustl.edu/gsc/cgi-bin/fpchuman.single.pl). BAC clones were obtained from Research Genetics.
QuMA.
PCR primer sequences for the microsatellite loci were obtained either from GDB and modified slightly or were designed in our lab (see Table 2, which is published as supporting information on the PNAS web site). The reference pool contained primer pairs for six different microsatellite loci located at different sites in the genome, which rarely showed alterations based on our previous CGH analysis of OSCC/SCCHN cell lines (C. M. Lese and S.M.G., unpublished data). The microsatellite markers used for the reference pool included D4S1605, D5S478, D12S1699, D14S988, D21S1904, and D22S922. All primer pairs were tested for PCR efficiency, and those with >90% efficiency were validated for QuMA by analysis of 12 unrelated anonymous control DNA samples from healthy individuals. QuMA was performed as described (16) by using 2 ng of genomic DNA for each PCR. The TaqMan CA-repeat fluorogenic probe used for all loci consisted of the following sequence: 5′-FAM (6-carboxy fluorescein)-TGTGTGTGTGTGTGTGTGTGT-TAMRA (6-carboxytetramethylrhodamine)-3′. All of the probes and primers for QuMA were purchased from Integrated DNA Technologies (Coralville, IA).
Validation of QuMA Results.
The QuMA results for all loci shown in Fig. 1A were validated by adding different amounts of pooled, normal control human genomic DNA into the QuMA reaction and by using the reference pool data from the 2 ng input for normalization. In this way, we simulated 4, 10, and 30 copies of the 11q13 region and showed that all loci gave an accurate representation of copy number in this range.
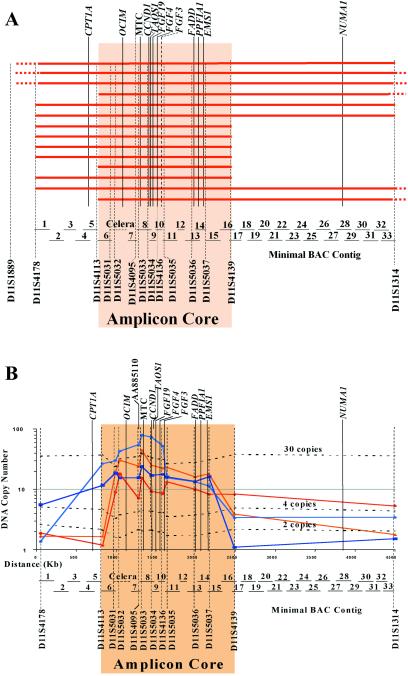
Fig 1.
Physical map of the 11q13 amplicon in oral cancer cell lines. (A) Region of amplification found in each of the 14 amplified cell lines. The red bars indicate the extent of amplification measured by QuMA for each cell line. Dashes at the end of a bar indicate that the amplified region extends outside of the area shown. The BAC contig is shown; a key to the names of the BACs can be found in Table 4, which is published as supporting information on the PNAS web site. (B) Map of the amplicon core showing the location of selected key genes in the amplicon and the QuMA data for five representative cell lines with amplification that define the amplicon core. Also shown are the locations of one new EST (AA885110) and the gene, TAOS1, both of which are highly amplified. The dashed lines represent the validation results of the primers used in QuMA. MTC, major translocation cluster identified in lymphomas.
Quantitative Reverse Transcription (RT)-PCR.
Taqman primers and probes were designed with the PRIMER EXPRESS V.2.0.0 (Applied Biosystems). Two separate reverse transcriptions were carried out with random hexamers by using a described protocol (19) and two RNA inputs (1 μg and 250 ng). No-reverse transcriptase controls were carried out for the highest RNA input each time. Quantitative PCR (QPCR) was performed on the cDNA on the ABI 7700 Sequence Detection Instrument (Applied Biosystems) and analyzed by using the described relative quantitation method (20). QPCR was performed for TAOS1, OCIM, CCND1, and ribosomal 18S RNA (endogenous control) with primers and probes listed in Table 3, which is published as supporting information on the PNAS web site. For the QPCR, the final concentrations of the reaction components were as follows: 1× PCR buffer A (Applied Biosystems), 300 nM each dNTP, 3.5 mM MgCl2, 0.06 units/μl Amplitaq Gold (Applied Biosystems), 500 nM (450 nM for CCND1 and 100 nM for 18S RNA) primers, and 200 nM (100 nM for CCND1 and 18S RNA) probe. The thermocycler conditions were 95°C Taq activation for 12 min and 45 cycles of 95°C denaturation for 15 s followed by 64°C anneal/extend (60°C for CCND1 and 18S RNA) for 60 s.
Results
Determination of the 11q13 Amplicon Core by QuMA.
Genomic DNA from 30 OSCC cell lines was analyzed by QuMA by using 19 (CA)n microsatellite markers that spanned more than 32 cM of chromosome 11q, including band 11q13. Of the cell lines examined, 19 (63%) had 11q13 amplification (data not shown), and 14 of these were amplified in the region between sequence-tagged site (STS) markers D11S4178 and D11S1314. Because the CCND1 gene and the t(11;14)(q13;q32) major translocation cluster (MTC) in mantle-cell lymphoma are located in this region, these 14 cell lines were examined further.
Ten microsatellite markers were chosen between D11S4178 and D11S1314 based on searching the raw genomic sequences, of which seven were used to fine-map the region (D11S5031–5037). Fig. 1A shows the results of QuMA analysis for 11q13 genomic amplification in the 14 OSCC cell lines. Amplicon size varied in different cell lines, but the minimal critical region shared by all 14 cell lines mapped between D11S4113 and D11S4139. The size of the 11q13 amplicon core was ≈1.5 Mb (Fig. 1B), and as expected, the CCND1 gene is located in the center along with several other well known genes. The copy number in all but one of the 14 amplified cell lines ranges from 10 to 30, which is consistent with the results of other studies (2). The only outlier has 79 copies of the amplified region (Fig. 1B). The extremely high copy number of the 11q13 amplicon in this cell line suggests that gene(s) in the amplicon drove the amplification process because they altered the critical homeostatic balance of the cells and provided an essential proliferative advantage, leading to tumor development.
Validation of the Accuracy of the QuMA Results.
The QuMA results were validated by adding 2, 8, and 28 copies of pooled genomic DNA from 5 unrelated healthy individuals into the QuMA experiment for each of the STS primer sets in the amplicon core, resulting in final values of 4, 10, and 30 copies. As expected, the copy number of each STS locus increased along with the increased DNA copy number added, with only slight variations (Fig. 1B).
Identification of the 11q13 Amplicon Breakpoints.
In the 19 OSCC cell lines with 11q13 amplification examined in this study, the proximal breakpoints in 11 of 19 (58%) cell lines mapped to two adjacent breakpoint intervals (Fig. 1A and Fig. 5, which is published as supporting information on the PNAS web site): one between D11S4178 and D11S4113, ≈550 kb in length, and the other between D11S4113 and D11S5031, ≈160 kb in size. In 6 of the 19 (32%) cell lines, the distal breakpoint mapped to one interval, between D11S5037 and D11S4139, ≈250 kb in length (Fig. 1A and Fig. 5). Fig. 1B shows four representative OSCC cell lines with sharp copy number changes between the two adjacent STS markers at the boundaries flanking the amplicon core, clearly defining the location of the amplicon breakpoints.
Assembly of a 5.0-Mb Physical Map of 11q13.
Because the region between STS markers D11S4178 and D11S1314 seemed to be highly amplified, a virtual BAC contig was constructed. Genes, ESTs, and STS sequences located between D11S4178 and D11S1314 were used in a blast search against the HTGS and nonredundant nucleotide sequence databases to identify the corresponding BACs. Next, “virtual chromosome walking” was carried out by blast screening with the repeat-sequence-masked end sequences of the existing BACs against the HTGS and nonredundant nucleotide sequence databases again and identifying additional BACs located on either side of the original BAC. By repeating this process, the minimal BAC contig was constructed (Fig. 1). The region between BACs RPCI11–554A11 and CMB9–31O9 comprises the only gap remaining. Approximately 250 kb of DNA sequence covering this gap was obtained from the Celera Human Genome Database, but no BAC clone information is available for this segment. The sequence of the entire 5-Mb contig is available.
There are more than 30 (CA)n microsatellite markers and 23 known genes in this BAC contig. In addition, previously uncharacterized ESTs in this region, for which the corresponding genes remain as yet unknown, were identified.
Positional Candidate Gene Analysis by Northern Blotting.
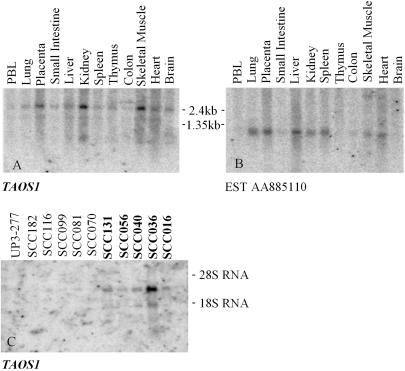
Exploration of the sequence data from the amplicon core revealed several previously uncharacterized EST clusters, suggesting the presence of unidentified genes. Two clusters mapped in close proximity to the CCND1 gene, and we chose to analyze them further. One EST from each cluster was selected to determine whether the putative gene was expressed in 12 normal tissues (CLONTECH MTN blot) and in OSCC cell lines. By using EST AI885296 (included in the TAOS1 gene) as a probe, the MTN blot showed a 2.5-kb main transcript and a 1.5-kb alternative minor transcript. TAOS1 was broadly expressed in all of the 12 tissues, with higher levels in placenta, kidney, and skeletal muscle (Fig. 2A). Another EST, AA885110 showed a ≈1-kb transcript that had little or no expression in brain, colon, thymus, small intestine, and peripheral blood leukocytes but had variable expression levels in the remainder of the tissues (Fig. 2B). In the Northern blot of the OSCC cell lines, AA885110 was not expressed (data not shown), but AI885296 (TAOS1) showed increased expression in the amplified cell lines, with low expression in the nonamplified cell lines and cultured normal human oral keratinocytes, serving as controls (Fig. 2C).
Fig 2.
Expression study of the new ESTs in the core of 11q13 amplicon. (A and B) Multiple tissue Northern blots for the two ESTs in the core of the 11q13 amplicon. EST AI885296 is included in TAOS1. Both show wide tissue expression. (C) EST AI885296/TAOS1 is expressed in oral cancer cell lines with 11q13 amplification (UPCI:SCC131, UPCI:SCC056, UPCI:SCC040, UPCI:SCC036, and UPCI:SCC016) and at much lower levels in cell lines without amplification and in normal human oral keratinocytes (UP3–277).
Identification of the TAOS1 Gene.
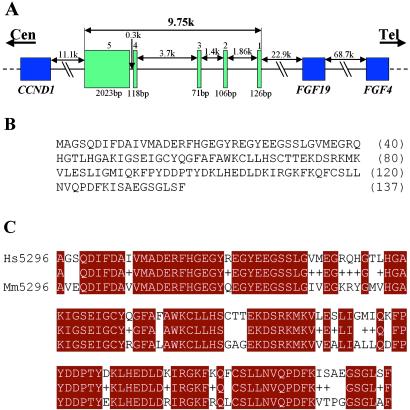
By using the sequence of EST AI885296, we searched the nonredundant nucleotide sequence database and found two Mammalian Gene Collection (MGC) clones, BE410345 (I.M.A.G.E. Consortium CloneID 3637085) and BG282549 (I.M.A.G.E. Consortium CloneID 4544931). Analysis after 5′ rapid amplification of cDNA ends (RACE) showed that the MGC clones contained all but 7 bp of the full length of the TAOS1 gene. After sequencing both clones and considering the 5′ RACE result, we concluded that TAOS1 spans 2,494 bp, which corresponded to our 2.5-kb Northern blot band. TAOS1 consists of five predicted exons and spans 9.75 kb of genomic DNA (Fig. 3A). According to our BLAST search results, TAOS1 corresponds to more than 50 ESTs in this EST cluster, and we believe that the minor 1.5-kb transcript seen on the Northern blots can be explained by alternate splicing of the exons encoding the 2.5-kb transcript. The exact sequence and structure of this minor transcript, however, remains to be determined. There are 137 amino acids in the protein sequence encoded by the 2.5-kb transcript, and the predicted molecular weight of the protein is 15.4 kDa (Fig. 3B). After comparing the amino acid sequence of TAOS1 with the nonredundant protein database, we found that it matched a full-length mouse gene (AK008702) with 77% (106/137) identity, indicating a high degree of evolutionary conservation (Fig. 3C). The function of TAOS1 is currently unknown.
Fig 3.
Identification of the TAOS1 gene. (A) Gene structure of TAOS1. The five exons are labeled 1–5. The coding region is 411 bp and covers all five exons. (B) Amino acid sequence of TAOS1. (C) Sequence alignment of TAOS1 from human (Hs) and mouse (Mm). Black boxes with white letters highlight identical residues, and the “+” signs indicate the conserved residues.
Expression Analysis of the TAOS1, CCND1, and OCIM Genes by Quantitative RT-PCR.
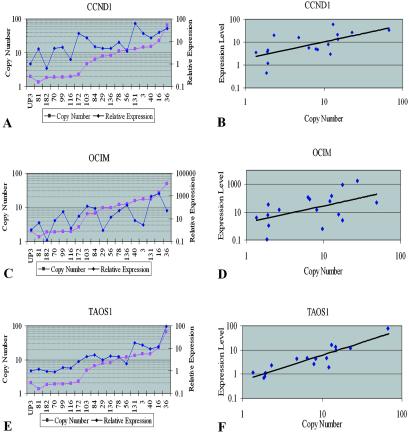
Previous studies of the 11q13 amplicon have suggested that CCND1 is the target gene of 11q13 amplification (5). Evidence also suggests that OCIM is a potential oncogene in multiple myeloma (15). To investigate the expression of TAOS1 compared with CCND1 and OCIM, we performed quantitative RT-PCR in 11 OSCC cell lines with 11q13 amplification and six OSCC cell lines without 11q13 amplification (Fig. 4). Normal keratinocytes were used as a control in this study. All 11 cell lines with 11q13 amplification showed increased expression of CCND1; four of six nonamplified cell lines also showed increased expression of CCND1 compared with the normal keratinocyte control (Fig. 4A).
Fig 4.
Expression of the CCND1, OCIM, and TAOS1 genes measured by TaqMan quantitative RT-PCR in the UPCI:SCC OSCC cell lines and the correlation between their gene copy number and gene expression. (A, C, and E) Expression levels of CCND1, OCIM, and TAOS1 in 17 OSCC cell lines; the first 6 cell lines in the graph do not have 11q13 amplification. Data are reported relative to 18S rRNA expression and normalized to normal human oral keratinocytes (ker). (B, D, and F) Correlation of gene copy numbers and expression levels.
In 14 of the 17 cell lines examined, OCIM showed elevated expression. However, one of the cell lines with 11q13 amplification, UPCI:SCC029, did not show increased expression of OCIM, and four of the six cell lines without 11q13 amplification showed increased expression. In two cell lines, UPCI:SCC131 and UPCI:SCC016, OCIM showed extreme overexpression compared with the normal keratinocyte control with 890- and 1,800-fold increases, respectively (Fig. 4C).
TAOS1 showed increased expression in all 11 cell lines with 11q13 amplification compared with a normal oral keratinocyte control (Fig. 4E). Overall, it is not overexpressed in cell lines without 11q13 amplification. Thus, TAOS1 gene copy number is more strongly correlated with its gene expression level (Fig. 4F) than are CCND1 or OCIM (Fig. 4 B and D).
Discussion
Numerous investigators have devoted significant effort toward elucidating amplicon structure and identifying potential oncogenes driving gene amplification at sites around the genome. In general, when an amplicon is identified by comparative genomic hybridization (CGH) in tumor cells, conventional methods such as Southern blot analysis are used to map the amplicon structure and estimate the relative copy number of the amplicon. Recently, the 17q22–23 amplicon was defined by Southern blot analysis in primary breast tumors and cell lines, and several independent gene targets of amplification were identified (21). However, Southern blot analysis is tedious, material consumptive, and relies on probe availability. Several other techniques, such as semiquantitative PCR (22, 23) and array CGH (24), have been used to circumvent the problems associated with amplicon assessment by Southern blotting. In the present study, the QuMA technique was used to map the 11q13 amplicon. QuMA requires very little DNA from precious samples, such as primary tumors, and is cost effective because it uses a single TaqMan probe for all test and reference loci. The completion of the human genome project greatly increased the availability of microsatellites for QuMA analysis, and, theoretically, the resolution of QuMA analysis could be as high as the distance between any two adjacent (CA)n microsatellite markers. Once a set of reference and test loci is prepared and validated, QuMA can be used to screen a large number of specimens in a short period. As our validation study showed, the estimation of DNA copy number by QuMA is precise, with only slight variations.
Chromosomal band 11q13 is one of the most frequently amplified genomic segments in tumors (2). Many groups have constructed physical maps of band 11q13 (10–15). In this study, a 5-Mb comprehensive physical map of the 11q13 amplicon was constructed, which includes the 1.5-Mb amplicon core. We found that 23 known genes reside in the 5-Mb region, and eight of them—OCIM, CCND1, FGF19, FGF3, FGF4, FADD, PPFIA1, and EMS1—are located in the amplicon core. In this study, the structure of the 11q13 amplicon has been finely mapped, and the copy number along the amplified region has been estimated precisely. It should be very useful for positional candidate cloning important genes and for further study of this region.
Investigators have proposed that gene amplification is caused by breakage-fusion-bridge (BFB) cycles (25, 26), with fragile sites being chromosomal hot spots predisposed to breakage during BFB cycles (27). Recently, this hypothesis was supported by drug-resistance gene amplification in Chinese hamster cells (28) and by MET oncogene amplification in human gastric carcinoma (29). From characterization of the 11q13 amplicon, we observed clustering of the proximal and distal amplicon breakpoints within distinct chromosomal regions. We had hypothesized that the proximal amplicon breakpoint may coincide with the major translocation cluster (MTC) seen in mantle cell lymphomas with the t(11;14)(q13;q32), but instead, the MTC was located in the middle of the 11q13 amplicon core. This finding suggests that different breakage mechanisms may be involved in these two processes. Our findings also suggest the presence of two or more as-yet-unidentified breakage hotspots or fragile sites that may be responsible for the breakpoints defining the 11q13 amplicon core. The observation of consistent breakpoints also supports our previous hypothesis that 11q13 amplification arises as a result of BFB cycles in OSCC (30).
Generally, it is believed that increased gene dosage by DNA amplification results in increased expression of a target gene, resulting in a selective advantage for the cells (1). The CCND1 gene has been widely considered to be the target gene of 11q13 gene amplification because of its overexpression and important role in driving the G1/S cell-cycle transition (5, 31). Earlier studies showed that besides CCND1, there are three other “independent” amplicons at 11q13 in breast cancer (32). One of these amplicons has been mapped in detail in breast cancer, and the amplicon core was narrowed to a 350-kb region encompassing D11S533 (12), which is located at 11q13.5-q14. None of the OSCC cell lines examined in this study was found to contain this amplicon. This finding suggests that the driving gene in the D11S533 amplicon may function in a different pathway important in the progress of breast cancer but not in OSCC. The other two “independent” amplicons, one marked by EMS1 and the other one, D11S97 (between D11S4113 and D11S5031 and approximately 500-kb centromeric to CCND1), have been shown to amplify independently of CCND1 in other tumor types (32–34). Evidence has highlighted the importance of the EMS1 gene as another possible target for 11q13 gene amplification (6, 35). Although no cell lines examined in our study show independent DNA amplification in these two regions, our results indicate that both EMS1 and D11S97 are included in the core of 11q13 amplicon in OSCC. Therefore, it is possible that in addition to CCND1 and EMS1, other genes may be important for driving the 11q13 amplification in OSCC.
The overexpression of CCND1 seen in the nonamplified OSCC cell lines by quantitative RT-PCR in this study suggests that increasing gene dosage is not the only means of up-regulating expression of the CCND1 gene. Other mechanisms seem to be causing increased CCND1 expression in the OSCC cells. In addition, the two cell lines with extreme overexpression of the OCIM gene are not the cell lines with the highest 11q13 amplicon copy number. The mixed expression profile among the cell lines with or without 11q13 amplification suggests that OCIM expression only partially depends on its copy number, and that there are also other mechanisms regulating its expression. Considering its ability to transform NIH 3T3 cells and its overexpression in some multiple myeloma cell lines with t(11;14)(q13;q32) (15, 36), OCIM may play a unique role in the progress of 11q13 amplification in a subset of OSCC and in other tumor types. This report quantifies OCIM expression in OSCC and in the context of 11q13 amplification.
By Northern blot and quantitative RT-PCR analyses, the TAOS1 gene, which is located approximately 12-kb distal to the CCND1 gene, is overexpressed in cell lines with 11q13 amplification, suggesting that this gene could be another important target gene for 11q13 gene amplification. The rearrangement and overexpression of CCND1 is a consistent feature of mantle cell lymphomas and leukemias associated with 11q13 translocations. In some cases, the rearrangement occurred in the 3′-UTR region of CCND1 (37, 38). Given the close proximity of TAOS1 to CCND1, it will be interesting to determine whether TAOS1 is rearranged and/or exhibits altered expression in these hematologic malignancies. The strong correlation between TAOS1 copy number and expression suggests that TAOS1 expression is copy number-dependent, and that TAOS1 could be another primary driving force behind 11q13 gene amplification along with the CCND1 gene. Although overexpression of CCND1 can occur in the absence of amplification in some HNSCC tumors, both amplification and overexpression have been linked to poor prognosis in this disease (39–41). As yet, we have not observed overexpression of TAOS1 in the absence of amplification, but only a relatively small number of cell lines have been examined compared with the vast literature available on CCND1. Furthermore, because of the tight correlation between expression and gene amplification, the expression status of TAOS1 may be a useful biomarker for the diagnosis and prognosis of OSCC. Further examination of the structure of the 11q13 amplicon in this and other tumor types and clarification of the function of TAOS1 and examination of its relationship with tumor growth and response to therapy should provide new insights into the mechanism behind and significance of gene amplification in cancer cells.
Supplementary Material
Acknowledgments
We thank Ms. Sarita Singh for culturing the NHOK cells and Dr. No-Hee Park for teaching us how to grow NHOK. We thank Ms. Elizabeth Lawrence in Dr. Robert Ferrell's laboratory for doing our sequencing at the high throughput sequencing facility in the University of Pittsburgh Center for Human Genetics and Integrative Biology. We thank Mr. Conover Talbot from Genome Database at Johns Hopkins University for naming the microsatellites and for helpful discussions. We thank Dr. David Ginzinger for providing the CCND1 primers and probe, and Drs. William Saunders and Richard Wood and Ms. Shalini Skarja for helpful discussion of the manuscript. This work was supported in part by National Institutes of Health Grants R01DE12008 (to S.M.G.) and P60DE13059, and the Oral Cancer Center at the University of Pittsburgh (to Dr. Eugene N. Myers).
Abbreviations
OSCC, oral squamous cell carcinomas
QuMA, quantitative microsatellite analysis
EST, expressed sequence tag
BAC, bacteria artificial chromosome
HTGS, high throughput genomic sequences
RT-PCR, reverse transcription–PCR
STS, sequence-tagged site
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF503940) and in the Genome Database (accession no. D11S5031–5037).
References
- 1.Savelyeva L. & Schwab, M. (2001) Cancer Lett. 167, 115-123. [DOI] [PubMed] [Google Scholar]
- 2.Schwab M. (1998) BioEssays 20, 473-479. [DOI] [PubMed] [Google Scholar]
- 3.Åkervall J. A., Jin, Y., Wennerberg, J. P., Zatterstrom, U. K., Kjellen, E., Mertens, F., Willen, R., Mandahl, N., Heim, S. & Mitelman, F. (1995) Cancer 76, 853-859. [DOI] [PubMed] [Google Scholar]
- 4.Gaudray P., Szepetowski, P., Escot, C., Birnbaum, D. & Theillet, C. (1992) Mutat. Res. 276, 317-328. [DOI] [PubMed] [Google Scholar]
- 5.Dickson C., Fantl, V., Gillett, C., Brookes, S., Bartek, J., Smith, R., Fisher, C., Barnes, D. & Peters, G. (1995) Cancer Lett. 90, 43-50. [DOI] [PubMed] [Google Scholar]
- 6.Schuuring E. (1995) Gene 159, 83-96. [DOI] [PubMed] [Google Scholar]
- 7.Gollin S. M. (2001) Head Neck 23, 238-253. [DOI] [PubMed] [Google Scholar]
- 8.Lese C. M., Rossie, K. M., Appel, B. N., Reddy, J. K., Johnson, J. T., Myers, E. N. & Gollin, S. M. (1995) Genes Chromosomes Cancer 12, 288-295. [DOI] [PubMed] [Google Scholar]
- 9.Jin Y., Höglund, M., Jin, C., Martins, C., Wennerberg, J., Åkervall, J., Mandahl, N., Mitelman, F. & Mertens, F. (1998) Genes Chromosomes Cancer 22, 312-320. [PubMed] [Google Scholar]
- 10.Tanigami A., Tokino, T., Takita, K., Takiguchi, S. & Nakamura, Y. (1992) Genomics 13, 16-20. [DOI] [PubMed] [Google Scholar]
- 11.Szepetowski P., Perucca-Lostanlen, D. & Gaudray, P. (1993) Genomics 16, 745-750. [DOI] [PubMed] [Google Scholar]
- 12.Bekri S., Adelaide, J., Merscher, S., Grosgeorge, J., Caroli-Bosc, F., Perucca-Lostanlen, D., Kelley, P. M., Pebusque, M. J., Theillet, C., Birnbaum, D. & Gaudray, P. (1997) Cytogenet. Cell Genet. 79, 125-131. [DOI] [PubMed] [Google Scholar]
- 13.Courseaux A., Szepetowski, P., Fernandes, M., Serizet, C., Kawaguchi, Y., Grosgeorge, J., Perucca-Lostanlen, D., Shows, T. B., Todd, J. A., Nowak, N. J. & Gaudray, P. (1997) Genomics 40, 13-23. [DOI] [PubMed] [Google Scholar]
- 14.Merscher S., Bekri, S., de Leeuw, B., Pedeutour, F., Grosgeorge, J., Shows, T. B., Mullenbach, R., Le Paslier, D., Nowak, N. J. & Gaudray, P. (1997) Genomics 39, 340-347. [DOI] [PubMed] [Google Scholar]
- 15.Janssen J. W., Vaandrager, J. W., Heuser, T., Jauch, A., Kluin, P. M., Geelen, E., Bergsagel, P. L., Kuehl, W. M., Drexler, H. G., Otsuki, T., et al. (2000) Blood 95, 2691-2698. [PubMed] [Google Scholar]
- 16.Ginzinger D. G., Godfrey, T. E., Nigro, J., Moore, D. H., 2nd, Suzuki, S., Pallavicini, M. G., Gray, J. W. & Jensen, R. H. (2000) Cancer Res. 60, 5405-5409. [PubMed] [Google Scholar]
- 17.Park N.-H., Min, B. M., Li, S.-L., Huang, M. Z., Cherick, H. M. & Doniger, J. (1991) Carcinogenesis 12, 1627-1631. [DOI] [PubMed] [Google Scholar]
- 18.Lennon G., Auffray, C., Polymeropoulos, M. & Soares, M. B. (1996) Genomics 33, 151-152. [DOI] [PubMed] [Google Scholar]
- 19.Godfrey T. E., Kim, S. H., Chavira, M., Ruff, D. W., Warren, R. S., Gray, J. W. & Jensen, R. H. (2000) J. Mol. Diagn. 2, 84-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perkin–Elmer Applied Biosystems, (1997) User Bulletin No. 2..
- 21.Wu G., Sinclair, C., Hinson, S., Ingle, J. N., Roche, P. C. & Couch, F. J. (2001) Cancer Res. 61, 4951-4955. [PubMed] [Google Scholar]
- 22.Lin L., Prescott, M. S., Zhu, Z., Singh, P., Chun, S. Y., Kuick, R. D., Hanash, S. M., Orringer, M. B., Glover, T. W. & Beer, D. G. (2000) Cancer Res. 60, 7021-7027. [PubMed] [Google Scholar]
- 23.Lin L., Aggarwal, S., Glover, T. W., Orringer, M. B., Hanash, S. & Beer, D. G. (2000) Cancer Res. 60, 1341-1347. [PubMed] [Google Scholar]
- 24.Albertson D. G., Ylstra, B., Segraves, R., Collins, C., Dairkee, S. H., Kowbel, D., Kuo, W. L., Gray, J. W. & Pinkel, D. (2000) Nat. Genet. 25, 144-146. [DOI] [PubMed] [Google Scholar]
- 25.Toledo F., Le Roscouet, D., Buttin, G. & Debatisse, M. (1992) EMBO J. 11, 2665-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma C., Martin, S., Trask, B. & Hamlin, J. L. (1993) Genes Dev. 7, 605-620. [DOI] [PubMed] [Google Scholar]
- 27.Kuo M. T., Vyas, R. C., Jiang, L. X. & Hittelman, W. N. (1994) Mol. Cell. Biol. 14, 5202-5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coquelle A., Pipiras, E., Toledo, F., Buttin, G. & Debatisse, M. (1997) Cell 89, 215-225. [DOI] [PubMed] [Google Scholar]
- 29.Hellman A., Zlotorynski, E., Scherer, S. W., Cheung, J., Vincent, J. B., Smith, D. I., Trakhtenbrot, L. & Kerem, B. (2002) Cancer Cell 1, 89-97. [DOI] [PubMed] [Google Scholar]
- 30.Shuster M. I., Han, L., Le Beau, M. M., Davis, E., Sawicki, M., Lese, C. M., Park, N. H., Colicelli, J. & Gollin, S. M. (2000) Genes Chromosomes Cancer 28, 153-163. [PubMed] [Google Scholar]
- 31.Wang M. B., Billings, K. R., Venkatesan, N., Hall, F. L. & Srivatsan, E. S. (1998) Otolaryngol. Head Neck Surg. 119, 593-599. [DOI] [PubMed] [Google Scholar]
- 32.Karlseder J., Zeillinger, R., Schneeberger, C., Czerwenka, K., Speiser, P., Kubista, E., Birnbaum, D., Gaudray, P. & Theillet, C. (1994) Genes Chromosomes Cancer 9, 42-48. [DOI] [PubMed] [Google Scholar]
- 33.Proctor A. J., Coombs, L. M., Cairns, J. P. & Knowles, M. A. (1991) Oncogene 6, 789-795. [PubMed] [Google Scholar]
- 34.Szepetowski P., Courseaux, A., Carle, G. F., Theillet, C. & Gaudray, P. (1992) Oncogene 7, 751-755. [PubMed] [Google Scholar]
- 35.Hui R., Campbell, D. H., Lee, C. S., McCaul, K., Horsfall, D. J., Musgrove, E. A., Daly, R. J., Seshadri, R. & Sutherland, R. L. (1997) Oncogene 15, 1617-1623. [DOI] [PubMed] [Google Scholar]
- 36.Janssen J. W., Braunger, J., Ballas, K., Faust, M., Siebers, U., Steenvoorden, A. C. & Bartram, C. R. (1999) Int. J. Cancer 80, 857-862. [DOI] [PubMed] [Google Scholar]
- 37.Rimokh R., Berger, F., Bastard, C., Klein, B., French, M., Archimbaud, E., Rouault, J. P., Santa Lucia, B., Duret, L. & Vuillaume, M. (1994) Blood 83, 3689-3696. [PubMed] [Google Scholar]
- 38.de Boer C. J., Vaandrager, J. W., van Krieken, J. H., Holmes, Z., Kluin, P. M. & Schuuring, E. (1997) Oncogene 15, 1599-1603. [DOI] [PubMed] [Google Scholar]
- 39.Michalides R., van Veelen, N., Hart, A., Loftus, B., Wientjens, E. & Balm, A. (1995) Cancer Res. 55, 975-978. [PubMed] [Google Scholar]
- 40.Meredith S. D., Levine, P. A., Burns, J. A., Gaffey, M. J., Boyd, J. C., Weiss, L. M., Erickson, N. L. & Williams, M. E. (1995) Arch. Otolaryngol. Head Neck Surg. 121, 790-794. [DOI] [PubMed] [Google Scholar]
- 41.Bockmuhl U., Kuchler, I. & Petersen, I. (2000) HNO 48, 451-456. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.