Abstract
Multiple pathways are responsible for transducing mechanical and hormonal stimuli into changes in gene expression during heart failure. In this study our goals were (i) to develop a sound statistical method to establish a comprehensive cutoff point for identification of differentially expressed genes, (ii) to identify a gene expression fingerprint for heart failure, (iii) to attempt to distinguish different etiologies of heart failure by their gene expression fingerprint, and (iv) to identify gene clusters that show coordinated up- or down-regulation in human heart failure. We used oligonucleotide microarrays to profile seven nonfailing (NF) and eight failing (F) human hearts with a diagnosis of end-stage dilated cardiomyopathy. Biological and experimental variability of the hybridization data were analyzed, and then a statistical analysis procedure was developed, including Student's t test after log-transformation and Wilcoxon Mann–Whitney test. A comprehensive cutoff point composed of fold change, average difference, and absolute call was then established and validated by TaqMan PCR. Of 6,606 genes on the GeneChip, 103 genes in 10 functional groups were differentially expressed between F and NF hearts. A dendrogram identified a gene expression fingerprint of F and NF hearts and also distinguished two F hearts with distinct etiologies (familial and alcoholic cardiomyopathy, respectively) with different expression patterns. K means clustering also revealed two potentially novel pathways associated with up-regulation of atrial natriuretic factor and brain natriuretic peptide and with increased expression of extracellular matrix proteins. Gene expression fingerprints may be useful indicators of heart failure etiologies.
Regardless of etiology, underlying mechanisms that contribute to development and progression of heart failure result from altered neurohormonal or mechanical (hemodynamic) stimuli (1). However, multiple molecular pathways are believed responsible for transducing these stimuli into changes in gene expression during the process of heart failure. Microarrays can provide an effective platform to examine these pathways in terms of mRNA expression (2).
Attempts using oligonucleotide (2) and cDNA (3) arrays have previously been made to identify genes differentially expressed in failing (F) and nonfailing (NF) human hearts. In a previous study from our laboratory (2), two NF donor hearts and two F human hearts with diagnoses of end-stage ischemic and dilated cardiomyopathy (DCM), respectively, were studied, and 19 genes were reported to be differentially expressed. Using a cardiovascular specific cDNA microarray, Barrans and colleagues examined two normal adult and two human hearts with diagnosis of hypertrophic cardiomyopathy. They reported 39 differentially expressed genes or expressed sequence tags (3). In this study, fold change in expression was used as the cutoff point.
The existence of significant biological variability in expression levels of many genes between human hearts is widely recognized. Experimental variability inherent in the microarray technology also exists. Consequently, many genes will show fairly large changes in gene expression by chance alone because of biological and/or experimental variability. Therefore, to correctly interpret microarray data, it is essential to include multiple samples in each group and to employ statistical methods capable of distinguishing chance occurrences from biologically meaningful data (4). Moreover, an evaluation of fold change without also considering absolute expression levels can be misleading in the interpretation of microarray data.
Therefore, we set three goals for this study: to establish sound statistical protocols resulting in a comprehensive cutoff point (including parameters for evaluation for both relative changes and absolute expression levels) to identify differentially expressed genes; second, to generate a gene expression profile of F human hearts from a large sample of F and NF hearts than previously attempted; and third, to determine whether heart failure of different etiologies is distinguished by distinct gene fingerprints.
Materials and Methods
mRNA Preparation and Hybridization to Oligonucleotide Array.
Tissue from left ventricular free wall was obtained from explanted F hearts of transplant recipients at the Cleveland Clinic Foundation (CCF) with a diagnosis of DCM. Tissue from NF hearts was from unmatched organ donors through Lifebanc of Northeast Ohio. All protocols were approved by the CCF Institutional Review Board and all patients gave informed consent. Age, medication, and an index of cardiac function (left ventricular ejection fraction) were obtained (Table 1). Tissue handling was as described (2, 5). Total RNA was isolated from the myocardium by using TRIzol reagent (GIBCO/BRL) as described by the manufacturer. Double-stranded cDNA was synthesized from 15 μg total RNA by using the Superscript Choice System (GIBCO/BRL) with an HPLC-purified oligo(dT) primer containing a T7 RNA polymerase promoter (GENSET, La Jolla, CA) following manufacturer's instructions. In vitro transcription was carried out with 1 μg cDNA using ENZO BioArray RNA transcript labeling kit (ENZO Diagnostics, Farmingdale, NY). Fragmentation of biotinylated cRNA (20 μg), protocols, and reagents for hybridization, washing, and staining followed instructions provided by Affymetrix.
Table 1.
Characteristics of nonfailing donor hearts and failing hearts
| Sample | Age | Sex | Diagnosis | LVEF | Treatment |
|---|---|---|---|---|---|
| Nonfailing | |||||
| NF1 | 26 | F | MVA | 70 | DP |
| NF2 | 38 | M | MVA | 70 | DP, EP, NE |
| NF3 | 52 | M | CVA | 45 | DP |
| NF4 | 29 | M | CVA | 50 | DZ, CD, EL, LP |
| NF5 | 69 | M | CVA | NA | DP, NE |
| NF6 | 46 | F | CVA | 70 | NE |
| NF8 | 52 | M | CVA | 85 | DP, TX |
| Failing | |||||
| F1 | 26 | M | DCM | 13 | DG |
| F2 | 49 | M | DCM | 20 | AD, CP, DG |
| F3 | 41 | M | DCM | 15 | AD, CP, DG, DT |
| F4 | 56 | M | DCM | 10 | AD, CP, DG, MN |
| F5 | 46 | M | DCM | 12 | AD, DG, LP, MN |
| F6 | 52 | M | DCM | 15 | CP, DG |
| F7 | 56 | M | DCM | 25 | AD, MN |
| F8 | 59 | M | DCM | 10 | AD, CP, DG, DT, MN |
LVEF, left ventricular ejection fraction; MVA, motor vehicle accident; CVA, cerebrovascular accident; DP, dopamine; EP, epinephrine; NE, norepinephrine; DZ, ditiazem; CD, clonidine; EL, enalapril; LP, lisinopril; TX, thyroxine; DG, digoxin; AD, amiodarone; CP, captopril; DT, dobutamine; MN, milrinone; LP, lisinopril.
TaqMan Quantitative Reverse Transcription (RT)-PCR.
We used the same total RNA pools for both microarray and RT-PCR analyses. RT of 5 μg total RNA was carried out in a 100-μl reaction using SuperScript kit (Life Technologies). Using 5 μl of the reverse reaction as template, quantitation was performed by using an ABI Prism 7700 sequence detection system (TaqMan) using the 5′ nuclease activity of TaqDNA polymerase to generate real time DNA analysis (7). Amplifications were generated by 10 min at 95°C and then 40 cycles of denaturation at 95°C for 15 s, annealing, and extension at 62°C for 1 min by using TaqMan universal PCR Master Mix kit (Applied Biosystems). TaqMan PCR for all four selected genes was performed by using 15 RNA specimens isolated from each of the 15 samples. The TaqMan assay was repeated either twice (connexin 43, cysteine protease, cAMP response element-binding protein) or three times (PLA2). To standardize the quantitation of four selected genes, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) from each sample was quantified on the same plate with the four target genes by using TaqMan GAPDH Control Reagents kit (Applied Biosystems).
Data Extraction and Statistical Analysis.
Detailed protocols for data extraction from Affymetrix oligonucleotide arrays and documentation on the sensitivity and quantitative aspects of the method have been described (2, 8). Affymetrix HuFl 6800 GeneChips containing 6,606 unique genes were used in this study with one GeneChip used for each human heart sample. To make comparisons across GeneChips, data sets on each GeneChip were normalized to a targeted total fluorescence of 300, representing total cRNA hybridized to the GeneChip. MICROSUITE 4.0 (Affymetrix) was used for data extraction.
Before any analyses were performed, all data were screened for any genes with a negative average difference value (negative values were considered as zero for further analysis). We used SAS/STAT 8.2 (SAS Institute, Cary, NC) for all statistical analyses. Tests for determination of normality and equal variances were performed to examine whether our data qualified for parametric statistical tests. Although data for the majority of genes were normally distributed (Shapiro–Wilk, P > 0.05) with equal variances among samples (equal variances test, P > 0.05), distribution of expression of levels of a number of genes did not satisfy criteria of normality (data not shown). Thus to use parametric analysis, a lognormal distribution was used. If g1F is gene 1 for an F heart, g1NF is the same gene for a NF heart, and x = ln g1F and y = ln g1NF, then g1F = ex and g1NF = ey, where x and y are normally distributed (data not shown), x and y can then be compared by using standard parametric methods, such as Student's t test. The average difference after log-transformation between F and NF hearts was compared by using Student's t test. GeneSpring (Silicon Genetics, Redwood City, CA) was used for data mining.
Results and Discussion
Reproducibility and Variability of Hybridization Data.
Before any statistical analysis was applied to the hybridization data, reproducibility of the data was assessed. The difference between the average percent of genes reported to be “present” (34% ± 4% vs. 34% ± 3%) and the mean average difference of the housekeeping gene GAPDH (27,329 ± 4,625 vs. 24,971 ± 2,181) between the NF and F groups, were not statistically significant (P > 0.05) (Table 4, which is published as supporting information on the PNAS web site, www.pnas.org).
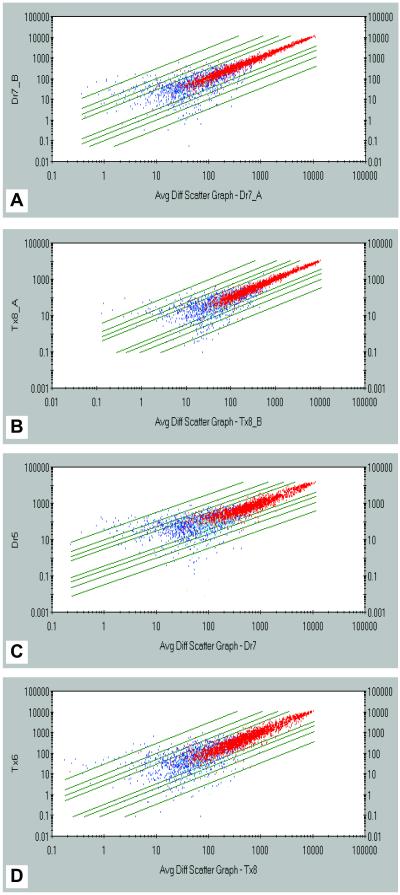
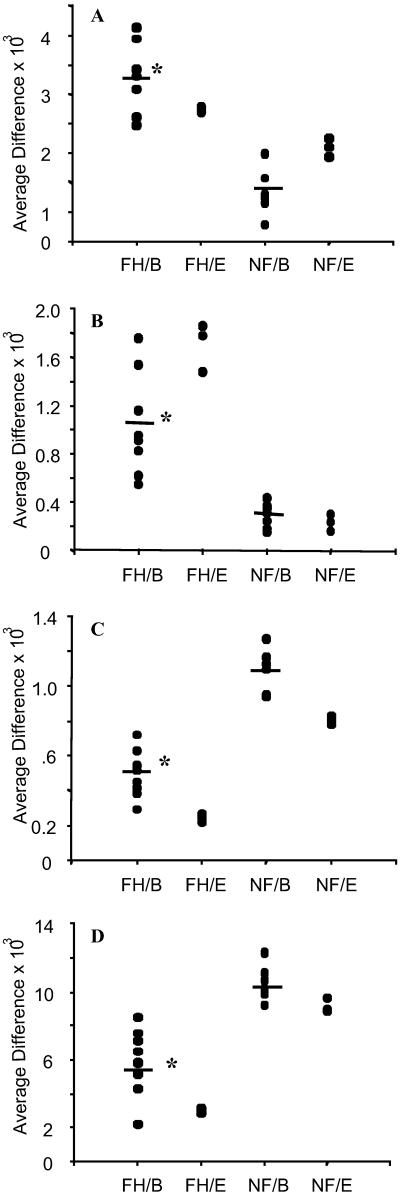
To identify major sources of experimental variability, two independent replicates of a sample of an F and NF heart, respectively, were performed. Overall expression patterns for the 6,606 genes on the Chip were plotted (Fig. 1). The expression levels of each of the genes for both the F and NF heart were highly correlated between two replicates for most of the genes on the Chip (Fig. 1 A and B). This was evident for genes called “present” (red dots) in both F and NF hearts and with an “average difference” > 150 units. However, expression levels for genes with a lower absolute intensity (average difference <150 units) and/or those identified as “absent” (blue dots) were not well correlated, indicating that the reported hybridization data from genes with low expression level and/or genes with an “absent” call are unreliable. Four examples of genes with high absolute intensity and a “present” call are shown in Fig. 2. These data indicate low experimental variability and high reproducibility.
Fig 1.
Scatter plots of the absolute intensity values for the 6,606 unique genes represented on the GeneChip. Red dots represent genes called present; yellow dots represent marginally present; blue dots represent absent; green lines (from inside to outside) represent 2-fold, 3-fold, 5-fold, and 10-fold change, respectively. (A) Experimental variability between two preps from the same NF (Dr) heart. (B) Experimental variability between two preps from the same F (Tx) heart. (C) Biological variability between two F hearts. (D) Biological variability between two NF hearts.
Fig 2.
Biological and experimental variability for four sample genes. *, P < 0.01, Student's t test after log-transformation and Wilcoxon test. FH/B, biological variability in eight F hearts; FH/E, experimental variability in three replicates of one F heart; NF/B, biological variability among seven NF hearts; NF/E, experimental variability in three replicates of one NF heart. (A) μ-Crystallin. (B) Thrombospondin-4. (C) α-Antichymotrypsin. (D) Apoliprotein D.
To establish a statistical protocol for data analysis, we also examined biological variability in gene expression between different hearts within the F and NF groups. We first compared the overall expression patterns for the 6,606 genes on the GeneChip between two samples. Although expression levels of most genes (red dots) between two samples in both NF and F groups were highly correlated (Fig. 1 C and D), there was noticeably more scatter of the data, as compared with the replicates (Fig. 1 A and B). This was evident when we plotted four examples (Fig. 2), thus indicating that biological variability is relatively high. These results also indicated that the variability (both biological and experimental) observed in the microarray data could be sufficiently high to generate a significant number of false positives if multiple samples are not included. It is also important to note that, as shown for experimental variability, both the absolute call and minimum absolute intensity need to be integrated into the cutoff point to minimize the rate of false positives.
Development and Implication of Statistical Protocols and a Comprehensive Cut-Off Point for Statistical Significance.
To minimize likelihood of false positives, we performed two independent analyses using parametric and nonparametric tests for genes that were potentially differentially expressed. Using a P < 0.01, corresponding to a concordance of 86% (9), the Wilcoxon Mann–Whitney test generated 371 potentially differentially expressed genes between NF and F hearts (Fig. 3). Although a high concordance value, in combination with fold-change, has been reported to significantly lower detection of false positives (9), the Wilcoxon test only takes into consideration absolute difference between two groups in generating a concordance value. Student's t test considers both absolute difference and its variance. Our first potential list of 229 genes was generated by using P < 0.01 from both Wilcoxon and Student's t tests (Fig. 3). The final list of 103 genes was obtained after implementation of our comprehensive cutoff points. A flow chart demonstrating the statistical approach used is shown in Fig. 3.
Fig 3.
Flowchart of steps for implementation of statistical protocols and our comprehensive cutoff points for data mining. 229 is the subset of 337 (Wilcoxon test, P < 0.01) and 270 (Student's t test, P < 0.01) that meet the criteria of P < 0.01 of both tests. Genes were filtered with mean of average difference > 200 units of the F heart group for up-regulated genes, and NF group for down-regulated genes. Genes were further filtered by absolute call = P or M in at least six of eight F hearts for up-regulated genes or five of seven NF hearts for down-regulated genes.
Validation by TaqMan Quantitative PCR.
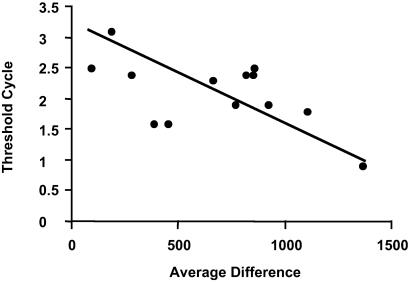
To validate our GeneChip data, we used TaqMan quantitative PCR on three genes with a fold change (1.7–1.8×, see Table 5, which is published as supporting information on the PNAS web site) close to one criterion (1.7×) of our comprehensive cutoff and one gene (PLA2) with higher fold change (5.1×, Table 2). All four genes, phospholipase A2, protease, connexin 43, and cAMP response element binding protein showed consistent changes with Taqman quantitative PCR and GeneChip analysis (Table 3). The relationship between cycle number (by Taqman PCR) and average difference (GeneChip) for one of these genes (Fig. 4) further validates our hybridization data.
Table 2.
Genes with most significant changes in expression between F and NF hearts
| GenBank accession no. | Name | Mean of NF, average difference units ±SD | Mean of F, average difference units ±SD | Fold change |
|---|---|---|---|---|
| Up-regulated genes | ||||
| M31776 | BNP | 751 ± 367 | 5,956 ± 1,908 | ↑ 7.9 |
| M54951 | ANF | 1,477 ± 1,228 | 6,249 ± 1,434 | ↑ 4.2 |
| M25296 | ANF precursor | 2,108 ± 751 | 6,986 ± 1,137 | ↑ 3.3 |
| M55998 | α1 collagen type I | 716 ± 319 | 2,667 ± 1,199 | ↑ 3.7 |
| Z74616 | Prepro-α2 collagen type I | 98 ± 79 | 485 ± 312 | ↑ 4.9 |
| D13666 | Osteoblast specific factor 2 | 40 ± 39 | 474 ± 281 | ↑ 12 |
| U21128 | Lumican | 462 ± 158 | 1,753 ± 561 | ↑ 3.8 |
| X06700 | Pro-α1 collagen type III | 156 ± 37 | 510 ± 229 | ↑ 3.3 |
| Z19585 | Thrombospondin-4 | 293 ± 101 | 1,040 ± 425 | ↑ 3.5 |
| M92934 | Connective tissue growth factor | 246 ± 110 | 794 ± 435 | ↑ 3.2 |
| Z24724 | Poly(A) site DNA | 196 ± 81 | 540 ± 114 | ↑ 2.7 |
| U10550 | GEM GTPase | 131 ± 29 | 359 ± 114 | ↑ 2.7 |
| M59287 | CDC-like kinase 1 | 126 ± 45 | 341 ± 84 | ↑ 2.7 |
| HG2755 | T-plastin | 87 ± 54 | 234 ± 98 | ↑ 2.7 |
| L02950 | Mμ-crystallin | 1,327 ± 372 | 3,389 ± 648 | ↑ 2.6 |
| Down-regulated genes | ||||
| M22430 | Phospholipase A2 | 1,563 ± 673 | 304 ± 92 | ↓ 5.1 |
| L19267 | Myotonin protein kinase | 1,072 ± 709 | 217 ± 186 | ↓ 4.9 |
| K02765 | Complement component 3 | 1,071 ± 332 | 230 ± 110 | ↓ 4.6 |
| HG3945 | Phospholipid transfer protein | 409 ± 130 | 12 ± 203 | ↓ 35 |
| M26311 | Cystic fibrosis antigen | 294 ± 114 | 22 ± 68 | ↓ 13 |
| X07315 | Nuclear transport factor | 255 ± 127 | 32 ± 152 | ↓ 7.8 |
| M12963 | Alcohol dehydrogenase I | 723 ± 188 | 150 ± 239 | ↓ 4.8 |
| M33195 | Fc-epsilon-receptor γ-chain | 316 ± 64 | 67 ± 64 | ↓ 4.7 |
| S80437 | Fatty acid synthase | 259 ± 162 | 55 ± 108 | ↓ 4.6 |
| M14539 | Factor XIII subunit | 915 ± 493 | 208 ± 614 | ↓ 4.4 |
| M80359 | MAP/microtubule affinity regulating kinase 3 | 319 ± 72 | 96 ± 112 | ↓ 3.3 |
| U42031 | Progesterone receptor-associated immunophilin | 456 ± 167 | 169 ± 109 | ↓ 2.7 |
| D17408 | Calponin | 1931 ± 1337 | 726 ± 2825 | ↓ 2.7 |
Genes are ranked by fold change and expression level.
Table 3.
Changes of four selected genes validated by TaqMan quantitative PCR
| GenBank accession no. | Name | GeneChip, fold change | TaqMan, fold change |
|---|---|---|---|
| M22430 | Phospholipase A2 | ↓ 5.1 | ↓ 8.5 ± 0.4 |
| D55696 | Cysteine protease | ↑ 1.8 | ↑ 1.7 ± 0.2 |
| X52947 | Connexin 43 | ↑ 1.8 | ↑ 1.6 ± 0.3 |
| U31903 | cAMP response element-binding protein | ↑ 1.7 | ↑ 1.5 ± 0.2 |
TaqMan PCR for 4 selected genes was performed in 15 RNA specimens isolated from 15 hearts. All data (threshold cycle) were standardized by GAPDH. The fold change was relative to the NF group.
Fig 4.
Correlation of average difference units of cysteine protease detected by hybridization to the GeneChip and threshold cycle by TaqMan quantitative PCR. Threshold cycle number was standardized by GAPDH, which was detected at the same time as cysteine protease. A lower value of threshold cycle indicates a higher original expression level (average difference) of cysteine protease.
Genes Differentially Expressed Between F and NF Hearts.
Of the 103 genes found to be differentially expressed in F and NF hearts, 65, or 63.1%, were up-regulated and 38, or 36.8%, were down-regulated. All of these genes were divided into 10 functional groups under the headings biomarkers, myofibrillar, extracellular matrix/cytoskeletal, proteolysis/stress, metabolism, apoptosis/inflammatory, signal transduction, immune system, and genes of unknown function (Table 2).
A comparison of genes differentially expressed between F and NF hearts in this study and the 19 genes previously reported by our laboratory to be differentially expressed (2) shows the increased expression of atrial natriuretic factor (ANF), μ-crystallin, and α1-antichymotrypsin measured previously in F hearts to be confirmed in the current study. αB-crystallin, myosin light chain 2, β-actin, and SLIM1 showed consistent changes in both studies but were not included in the final list in this study because they did not meet our comprehensive cutoff point (Fig. 3)
In F hearts, ANF, ANF precursor, and brain natriuretic peptide (BNP) were ranked as the top three of all up-regulated genes in terms of fold change and expression level. We defined this subgroup as biomarkers of heart failure. Consistent with the fact that during development of heart failure, myocytes revert to a pattern of fetal gene expression, e.g., increased expression of myosin heavy chain-β (10, 11) and replacement of cardiac α-actin by skeletal α-actin (11), cardiac α-actin was found to be down-regulated and myosin heavy chain-β up-regulated in the F hearts (Table 5).
Genes encoding extracellular matrix/cytoskeletal proteins represent another group with a significant increase in expression in F hearts. The majority of these genes (77%, 10 of 13) were up-regulated (Table 5). Considering the maladaptive remodeling process that occurs in F hearts, this was not unexpected. Increased expression of collagen types I and III, the major collagen subtypes in the myocardium and major determinants of tissue stiffness, represents an adverse environment for cardiac myocytes and plays a central role in the progressive deterioration of F hearts (13, 14). Although most genes in this subset were previously reported to show increased expression in heart failure, changes in expression of fibromodulin, t-plastin, fibronectin, and desmosome-associated protein were not previously reported.
Consistent with the fact that proteolysis and activation of a stress response occurs in F hearts in response to neurohormonal and mechanical injury (15, 16), the majority (80%, 8 of 10) of genes encoding proteolysis/stress related proteins showed increased expression (Table 5). It is interesting to note a 3.5-fold up-regulation of thrombospondin-4 in F hearts. Similar findings were reported in two recent gene expression profiling studies in a rat myocardial infarct model (9) and in skeletal muscle from muscular dystrophy patients (6). Although the implications of thrombospondin-4 up-regulation in heart failure is unknown, association of thrombospondin-4 with cardiac disease is highlighted by a recent report indicating that a missense variant (A387P) is associated with premature myocardial infarction (17).
Seventeen genes encoding proteins involved in metabolism were differentially expressed (Table 5). It is interesting to note that genes encoding proteins involved in fatty acid metabolism, e.g., apoliprotein D, fatty acid synthase, phospholipid transfer protein, were exclusively down-regulated. However, genes related to glucose metabolism, such as fructose 1–6 biphosphatase and mitochondrial NADP(+)-dependent malic enzyme, were up-regulated. These observations may indicate the fact that the primary myocardial energy source switches from fatty acid to glucose in heart failure (18).
Metallothionein and metallothionein 1L are stress-inducible and metal binding proteins. Their antioxidant function and regulation of apoptosis in the heart has previously been reported (20). α1-antichymotrypsin, a serine protease inhibitor, has been reported to attenuate myocardial ischemic injury (21, 22). Consistent with observations of increased apoptosis in F human hearts (19), decreased expression of α1-antichymotrypsin (3), metallothionein, and metallothionein 1L, may indicate that the anti-apoptosis pathway is inhibited in F hearts.
Elevated local levels of transforming growth factor (TGF)-β1 are believed to be involved in triggering remodeling of the extracellular matrix of the myocardium in DCM (23). Up-regulation of two TGF-β binding proteins in F hearts with dilated cardiomyopathy may be related to local elevation of TGF-β1.
Clustering of Differentially Expressed Genes.
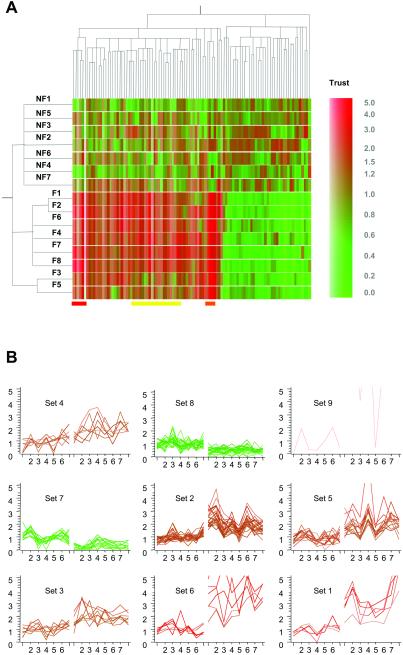
Distinct from clustering into functional groups, two unsupervised clustering algorithms were applied to the 103 differentially expressed genes. We used a hierarchical clustering method to generate a dendrogram for gene expression patterns across the 15 hearts. The results recapitulated the pattern of differences in gene expression between NF and F hearts (Fig. 5A). Of note, F hearts 3 and 5 were clustered together, away from the six other F hearts (Fig. 5A). Whereas all eight of these F hearts were diagnosed with DCM, F heart 3 was from a patient with alcoholic cardiomyopathy, and F heart 5 was from a patient with familial cardiomyopathy. Gene clusters associated with heart failure of a specific etiology are marked with a yellow bar (Fig. 5A). Genes encoding proteins involved in signal transduction or transcription, e.g., myocyte-specific enhancer factor 2, regulator of G protein signaling 2, stromal cell-derived factor 1, fibroblast growth factor 1, and also cardiac-specific genes such as myosin heavy chain β, connexin 43, fibromodulin and α2-actinin, were up-regulated in all of the other six F hearts, but only slightly up-regulated in F hearts 3 and 5 compared with the NF hearts. These are indicated as sets 2 and 5 in Fig. 5B. An expression profiling study on four different mouse cardiac hypertrophy models also demonstrated divergent transcriptional responses to independent genetic causes of cardiac hypertrophy (24). Findings from both F human hearts and mouse models indicated that heart failure of different etiology may involve different genetic determinants for their development. Further comparison of these two studies revealed a number of genes such as ANP, osteoblast specific factor 2, cytochrome P450, metallothionein 1, phospholipase A2, collagen, α2-actinin, etc., showing consistent changes in expression level in both studies.
Fig 5.
Hierarchical clustering of 15 hearts and 103 genes. (A Left) Dendrogram of 15 different heart samples, 7 NF and 8 F, recapitulates the major distinction between 7 NF and 8 F based on their similarity in genes expression. The total of 103 genes were clustered based on their expression patterns among 15 hearts. A trust color bar indicates the relative changes (fold) of mean absolute intensity of F group over NF group. (B) Nine gene clusters generated by K means (K = 9) clustering algorithm. Sets 1 and 6 correspond to the clusters of gene shown by red and orange bar, respectively (A). Most genes in set 5 and a partial list of genes in set 2 correspond to clusters of genes shown by the yellow bar (A). On the x axis, the first 7 hearts are NF hearts and the next 8 are F hearts.
A K means (K = 9) clustering algorithm was also applied to recluster the 103 genes based on their expression patterns among the 15 hearts. This method clusters genes that show coordinated fluctuations in gene expression across the 15 samples. The resulting nine clusters (Fig. 5B) were highly consistent compared with those generated by hierarchical clustering (data not shown). Based on the assumption that groups of genes that fluctuate together in expression levels may be regulated by common mechanisms, or alternatively, that one or more genes in the cluster regulates expression of other genes of the cluster, we attempted to interpret the gene expression patterns in some of these sets, in the context of possible molecular pathways involved in heart failure.
The most prominent clusters were sets 1 and 6, which showed significantly enhanced gene expression in F hearts (Fig. 5B). Three collagen isoforms, thrombospondin-4 and insulin-derived growth factor I (IGF-I) were clustered in set 1, defined as extracellular matrix related genes. It was not unexpected to see IGF-I in the same set as collagen because IGF-I up-regulation occurs in patients with hypertrophic cardiomyopathy (25). Furthermore, IGF-I overexpression of fibroblasts plays an important role in tissue remodeling during heart failure. IGF-I-stimulated fibroblast proliferation and myofibrillar collagen synthesis has been demonstrated in the rat heart (26). Another interesting set was set 6, defined as biomarkers for heart failure (ANF, BNP, ANF precursor, Lumican, and GEM GTPase). Of 103 genes, all three biomarkers for heart failure clustered in the same set. However, whether there is a link between GEM and ANF/BNP up-regulation remains to be determined. GEM GTPase is a mitogen-induced RAS-related GTP-binding protein overexpressed in skeletal muscle with type II diabetes mellitus (27, 28). Involvement of two mitogen activated Ras proteins in pathways for both ANF and BNP up-regulation in cardiac myocytes has been reported (29, 30). Whether increased expression of this Ras protein is involved in ANF and BNP up-regulation is an interesting future direction. Thus, unsupervised clustering of genes according to expression patterns across hearts permits identification of potentially coregulated genes or pathways regulating gene expression.
In summary, we conclude that the gene expression profile identified in this study represents a gene fingerprint for human heart failure and that unsupervised clustering segregates two failing hearts with different diagnoses, thus offering the exciting possibility of identifying DCM hearts of distinct etiologies.
Supplementary Material
Acknowledgments
We acknowledge the assistance of Dr. Jiacheng Yang and Carley Gwin. We thank Drs. Wilson Tang and Gary Francis for valuable discussions and Robin Lewis for help with manuscript preparation. We thank the Cardiac Transplant Team in the Department of Thoracic and Cardiovascular Surgery, residents in the Department of Pathology and Life Banc of Northeast Ohio. The work was supported by National Institutes of Health Grants R01 HL56256 and R01 AG16613 (to M.B.), Training Program in Vascular Cell Biology (T32 HL07914), F32 HL70496 (to F.-L.T.), the Kaufman Center for Heart Failure, and the Technology Transfer Fund and Department of Molecular Cardiology, Lerner Research Institute, Cleveland Clinic Foundation.
Abbreviations
NF, nonfailing
F, failing
DCM, dilated cardiomyopathy
BNP, brain natriuretic peptide
ANF, atrial natriuretic factor
GAPDH, glyceraldehyde-3-phosphate dehydrogenase
References
- 1.Mann D. L. (1999) Circulation 100, 999-1008. [DOI] [PubMed] [Google Scholar]
- 2.Yang J., Moravec, C. S., Sussman, M. A., DiPaola, N. R., Fu, D., Hawthorn, L., Mitchell, C. A., Young, J. B., Francis, G. S., McCarthy, P. M. & Bond, M. (2000) Circulation 102, 3046-3052. [DOI] [PubMed] [Google Scholar]
- 3.Barrans J. D., Stamatiou, D. & Liew, C. C. (2001) Biochem. Biophys. Res. Commun. 280, 964-969. [DOI] [PubMed] [Google Scholar]
- 4.Rao J. S. & Bond, M. (2001) Circ. Res. 88, 1226-1227. [DOI] [PubMed] [Google Scholar]
- 5.Zakhary D. R., Moravec, C. S., Stewart, R. W. & Bond, M. (1999) Circulation 99, 505-510. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y.-W., Zhao, P., Broup, P. & Hoffman, E. P. (2000) J. Cell Biol. 151, 1321-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heid C. A., Stevens, J., Livak, K. J. & Williams, P. M. (1996) Genome Res. 6, 986-994. [DOI] [PubMed] [Google Scholar]
- 8.Lipshitz R. J., Fodor, S. P., Gingerous, T. R. & Lockhart, D. J. (1999) Nat. Genet. 21, 20-24. [DOI] [PubMed] [Google Scholar]
- 9.Jin H. K., Yang, R. H., Awad, T. A, Wang, F., Li, W., Williams, S. P., Ogasawara, A., Shimada, B., Williams, M. & de Feo, G. (2001) Circulation 103, 736-742. [DOI] [PubMed] [Google Scholar]
- 10.Swynghedauw B. (1986) Physiol. Rev. 66, 710-771. [DOI] [PubMed] [Google Scholar]
- 11.Sugden P. H. & Fuller, S. J. (1997) in Heart Failure, eds. Poole-Wilson, P. A., Colucci, W. S., Massie, B. M., Chatterjee, K. & Coats, A. J. S. (Churchill Livingston, New York), pp. 47–48.
- 12.Maguchi M., Nishida, W., Kohara, K., Kuwano, A., Kondo, I. & Hiwada, K. (1995) Biochem. Biophys. Res. Commun. 217, 238-244. [DOI] [PubMed] [Google Scholar]
- 13.Schaper J., Froede, R., Hein, S., Buck, A., Hashizume, B. H., Speiser, B., Friedl, A. & Bleese, N. (1991) Circulation 83, 504-514. [DOI] [PubMed] [Google Scholar]
- 14.Masutomo K., Makino, N., Sugano, M., Miyamoto, S. & Yanaga, T. (1999) J. Mol. Cell. Cardiol. 31, 1607-1615. [DOI] [PubMed] [Google Scholar]
- 15.Margossian S. S., Anderson, P. A., Chantler, P. D., Deziel, M., Umeda, P. K., Patel, H., Stafford, W. F., Norton, P., Malhotra, A., Yang, F., et al. (1999) Mol. Cell. Biochem. 194, 301-313. [DOI] [PubMed] [Google Scholar]
- 16.Vatner S. F., Vatner, D. E. & Homcy, C. J. (2000) Circ. Res. 86, 502-506. [DOI] [PubMed] [Google Scholar]
- 17.Topol E. J., McCarthy, J., Gabriel, S., Moliterno, D. J., Rogers, W., Newby, K., Freedman, M., Metivier, J., Cannata, R., O'Donnell, C. J., et al. (2001) Circulation 104, 2641-2644. [DOI] [PubMed] [Google Scholar]
- 18.Sack M. N. & Kelly, D. P. (1998) Int. J. Mol. Med. 1, 17-24. [DOI] [PubMed] [Google Scholar]
- 19.Saraste A., Pulkki, K., Kallajoki, M., Heikkila, P., Laine, P., Mattila, S., Nieminen, M. S., Parvinen, M. & Voipio-Pulkki, L. M. (1999) Eur. J. Clin. Inv. 29, 380-386. [DOI] [PubMed] [Google Scholar]
- 20.Kang Y. J. (1999) Pro. Soc. Exp. Bio. Med. 222, 263-273. [DOI] [PubMed] [Google Scholar]
- 21.Murohara T., Guo, J. P. & Lefer, A. M. (1995) J. Pharmacol. Exp. Ther. 274, 1246-1253. [PubMed] [Google Scholar]
- 22.Delyani J. A., Murohara, T. & Lefer, A. M. (1996) Am. J. Physiol. 270, H881-H887. [DOI] [PubMed] [Google Scholar]
- 23.Bornstein P. & Sage, H. (1989) Prog. Nucleic Acid Res. Mol. Biol. 37, 67-106. [DOI] [PubMed] [Google Scholar]
- 24.Aronow B. J., Toyokawa, T., Canning, A., Haghighi, K., Delling, U., Kranias, E., Molkentin, J. D. & Dorn, G. W., II (2001) Physiol. Genomics 6, 19-28. [DOI] [PubMed] [Google Scholar]
- 25.Ren J., Samson, W. K. & Sowers, J. R. (1999) J. Mol. Cell. Cardiol. 31, 2049-2061. [DOI] [PubMed] [Google Scholar]
- 26.Reiss K., Cheng, W., Kajstura, J., Sonnenblick, E. H. & Meggs, L. G. (1995) Am. J. Physiol. 269, H943-H951. [DOI] [PubMed] [Google Scholar]
- 27.Maguire J., Santoro, T., Jensen, P., Siebenlist, U., Yewdell, J. & Kelly, K. (1994) Science 265, 241-244. [DOI] [PubMed] [Google Scholar]
- 28.Reynet C. & Kahn, C. R. (1993) Science 262, 1441-1444. [DOI] [PubMed] [Google Scholar]
- 29.He Q. & LaPointe, M. C. (1999) Hypertension 33, 283-289. [DOI] [PubMed] [Google Scholar]
- 30.Jette C. & Thorburn, A. (2000) FEBS Lett. 467, 1-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.