Abstract
A family of small, single-span membrane proteins (the FXYD family) has recently been defined based on their sequence and structural homology. Some members of this family have already been identified as tissue-specific regulators of Na,K-ATPase (NKA). In the present study, we demonstrate that phospholemman (PLM) (FXYD1), so far considered to be a heart- and muscle-specific channel or channel-regulating protein, associates specifically and stably with six different α-β isozymes of NKA after coexpression in Xenopus oocytes, and with α1–β, and less efficiently with α2–β isozymes, in native cardiac and skeletal muscles. Stoichiometric association of PLM with NKA occurs posttranslationally either in the Golgi or the plasma membrane. Interaction of PLM with NKA induces a small decrease in the external K+ affinity of α1–β1 and α2–β1 isozymes and a nearly 2-fold decrease in the internal Na+ affinity. In conclusion, this study demonstrates that PLM is a tissue-specific regulator of NKA that may play an essential role in muscle contractility.
The Na,K-ATPase (NKA) is an ubiquitous plasma membrane enzyme that transports 3 Na+ out and 2 K+ into the cells by using the energy of the hydrolysis of ATP. NKA activity permits the creation and the maintenance of Na+ and K+ gradients across cell membranes that are essential for both cellular and body ion homeostasis. In addition to this functional role, NKA plays a role as a receptor for cardiac glycosides widely used in the treatment of heart failure because of their positive ionotropic action (1) and presumably for endogenous digitalis-like compounds identified in mammals (2). The minimal functional unit of NKA comprises a catalytic α subunit containing the cation, ATP and phosphate binding sites, and a glycosylated β subunit required for the correct folding and functional maturation of the α subunit (3). Four α and 3 β isoforms, which may form different, tissue-specific NKA isozymes with distinct transport and pharmacological properties, have been identified (4, 5).
Regulation of NKA activity is an important and complex process that involves short- and long-term mechanisms. Short-term regulation of NKA is either mediated by changes in intracellular Na+ concentrations that directly affect the Na,K-pump activity or by peptide hormone-mediated phosphorylation/dephosphorylation reactions leading to changes in the Na,K-pump transport properties or in its cell surface expression. On the other hand, long-term regulation involves mineralocorticoid or thyroid hormone-mediated changes in the transcription of α- and/or β-subunit genes leading to an increased expression level of Na,K-pumps (6). Recently, a new type of regulation has emerged that involves the association of small, single-span membrane proteins with NKA. These proteins belong to the so-called FXYD family, the members of which share a common signature sequences encompassing the transmembrane domain and adjacent regions (7). Three of the seven members of the FXYD family have so far been identified as regulators of NKA. Two FXYD2 variants (the so-called γa and γb subunits of NKA) (7–10) and FXYD4 (CHIF) (11, 12) were shown to be renal, nephron segment-specific regulators (13–16), whereas FXYD7 is a brain- and isozyme-specific regulator of NKA (44). The functional characteristics of three other members of this family, FXYD3 (MAT-8) (17), FXYD5 (RIC) (18), and FXYD6 (phosphohippolin) (19), have not yet been studied. Finally, the last member of the FXYD family, FXYD1, originally named phospholemman (PLM) (20), has been extensively investigated, but its actual physiological role remains obscure. FXYD1 is widely distributed with highest expression in heart and skeletal muscle (21), where it is the main substrate for protein kinase A and C (20). Expression of FXYD1 in oocytes or addition of the purified protein to lipid bilayers produces a chloride-sensitive current slowly activated by hyperpolarization (22). Moreover, PLM is able to switch among different conformations having different selectivities for cations and anions that permits the translocation of zwitterionic molecules such as taurine (23). More recently, Mahmmoud et al. (24) reported that a PLM-like protein coimmunoprecipitated with NKA from shark rectal glands. In view of these results, we tested whether, similar to FXYD2, -4, and -7, PLM is also a tissue-specific regulator of NKA. We demonstrate that PLM is indeed able to associate with NKA after coexpression in Xenopus oocytes as well as in native cardiac and skeletal muscle. PLM induces a small decrease in the apparent K+ affinity, as well as a 2-fold decrease in the apparent Na+ affinity of NKA isozymes. The reduction in the apparent Na+ affinity of NKA may be of physiological relevance during muscle contraction.
Materials and Methods
cDNAs.
Dog PLM cDNA, subcloned between the 5′ and 3′ domains of Xenopus β-globin, was kindly provided by B. Attali (Weizmann Institute of Science). Cloning of human NKA α1, α2, α3, and β1 cDNAs has been described (4). cDNAs for rat α1, α2*, and α3* (*, ouabain-resistant; ref. 25) and β1 subunits were kindly provided by J. Lingrel (University of Cincinnati) and cDNAs for the human sarcoendoplasmic reticulum Ca2+-ATPase 2a (SERCA2a) by D. H. MacLennan (University of Toronto). All cDNAs were introduced into the pSD5 vector and cRNAs were prepared by in vitro translation (26).
Protein Expression in Xenopus laevis Oocytes.
Stage V–VI oocytes were obtained from X. laevis as described (27). cRNAs coding for dog PLM, rat NKA α1, α2*, and α3*, and β1 subunits and rat SERCA2a were injected into oocytes in different combinations as described in the figure legends. To study protein expression and association, oocytes were incubated in modified Barth's solution (MBS), either in the absence or presence of 5 μg/ml brefeldin A (27), which retains proteins in the endoplasmic reticulum, and in the presence of 0.4–0.5 mCi/ml (1 Ci = 37 GBq) [3H]leucine (Amersham Pharmacia). Oocytes were subjected to 6–24-h pulse and to 24–48-h chase periods in MBS containing 10 mM cold leucine. After various pulse–chase periods, microsomes were prepared as described (27) and subjected to immunoprecipitations with PLM- or α-antibodies under nondenaturing conditions (see below). For control of PLM expression, metabolically labeled microsomes were directly loaded on SDS/Tricine gels (prepared according to the manufacturer's instructions) and revealed by fluorography.
Preparation of Microsomes from Bovine Heart and Rat Skeletal Muscle and Kidney.
Microsomes from bovine or rat cardiac sarcolemma (28) and from rat kidney (13) were prepared as described. For the preparation of microsomes from skeletal muscle, 1–2 g of rat skeletal muscle was finely cut, frozen, and ground in a mortar. The tissue was homogenized with a Polytron in a homogenization buffer containing 30 mM histidine (pH 7.4), 5 mM EDTA, 0.2 mM phenylmethylsulfonyl fluoride (PMSF), and 250 mM sucrose. The homogenate was centrifuged twice at 7,000 × g for 15 min and the pooled supernatants were centrifuged at 48,000 × g for 1 h. Microsomes were resuspended in homogenization buffer.
Immunoprecipitation and Western Blotting.
Rat and human NKA isozymes and SERCA2a, expressed and metabolically labeled in oocytes, were immunoprecipitated as described (29). A polyclonal PLM-antibody was produced against a peptide CRSSIRRLSTRRR of the C terminus of rat PLM, as described for γa and γb (10), and was used to immunoprecipitate PLM expressed in oocytes under nondenaturing conditions (26). Immunoprecipitates were resolved on 5–13% SDS-polyacrylamide gels and revealed by fluorography.
Coimmunoprecipitation of PLM from bovine heart sarcolemma microsomes was done as described (12), using a specific monoclonal α1-antibody (6H; kindly provided by M. Caplan, Yale University, New Haven, CT) or an α2-antibody, raised against the peptide HERED (ref. 30; kindly provided by T. Pressley, Texas Tech University, Houston). Western blots were probed with the PLM-antibody or anti-KETYY, which recognizes the C terminus of α subunits.
To demonstrate association of PLM with NKA in rat skeletal muscle, 500 μg of microsomal protein were incubated in digitonin buffer (100 mM NaCl/0.5% digitonin/5 mM PMSF/20 mM Tris⋅HCl, pH 7.4) for 5 h at 4°C. After centrifugation (500 × g for 2 min), supernatants were incubated with PLM-antibodies overnight at 4°C. After addition of Sepharose A beads and several washes in the presence and absence of digitonin, immune complexes were migrated on an 8% SDS-polyacrylamide gel and transferred onto nitrocellulose. NKA α isoforms were revealed with α1 and α2 isoform-specific antibodies (ref. 30; kindly provided by T. Pressley) and chemiluminescence detection (ECL, Amersham Pharmacia).
Electrophysiology.
Electrophysiological measurements were performed 3 days after oocyte injection with rat α1–β1 or α2*–β1 cRNAs alone or together with PLM cRNA by using the two-electrode voltage clamp technique. Measurements of the apparent external K+ affinity were carried out in the presence of 1 μM ouabain, which inhibits the endogenous, oocyte Na,K-pumps, but not the expressed ouabain-resistant rat NKA isozymes, as described (13). The maximal Na,K-pump current and the apparent K+ affinity (K1/2K+) measured in the presence or absence of external Na+ were obtained by fitting the Hill equation to the data with Hill coefficients of 1.6 or 1, respectively (31). Measurements of the apparent Na+ affinity of NKA in intact cells were performed as described (32) by coexpressing rat α1 or α2* with rat β1 cRNAs together with rat epithelial Na+ channel (ENaC) α-, β-, and γ-subunit cRNAs in the presence or absence of PLM cRNA. The maximal current (Imax) and the apparent Na+ affinity (K1/2Na+) were fitted by using the Hill equation and a Hill coefficient of 3 (32).
Ouabain Binding Assay.
Three days after cRNA injection of PLM together with human α1–β1, α2–β1, or α3–β1 cRNAs, microsomes were prepared and equilibrium ouabain binding experiments were carried out as described (4). Briefly, freeze–thawed permeabilized microsomes were incubated at 37°C in a medium containing 4 mM ATP, 4 mM MgCl2, 100 mM NaCl, 30 mM imidazole⋅HCl (pH 7.4), and various [3H]ouabain concentrations (1 × 10−9 to 1 × 10−7 M). After incubation for 2 h, aliquots corresponding to 5 μg of protein were removed, rapidly filtered under vacuum on glass fiber filters (Whatman GF/C), and rinsed three times with 4 ml of an ice-cold buffer containing 100 mM NaCl and 30 mM imidazole⋅HCl (pH 7.4). Radioactivity bound to filters was counted after addition of 4 ml of scintillation solution (Emulsifior scintillator plus, Packard).
Results
Specific Association of PLM with NKA in Xenopus Oocytes.
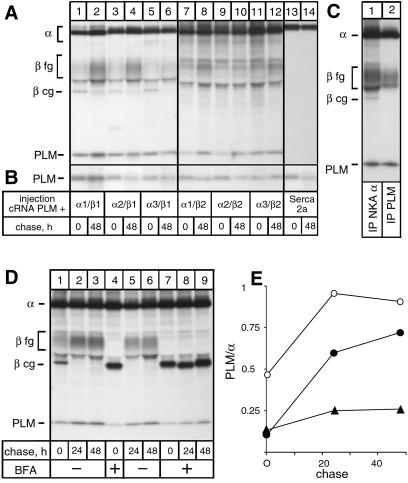
The specific association of PLM with NKA was investigated by coexpressing PLM in Xenopus oocytes together with rat NKA α1–β1, α2–β1, α3–β1, α1–β2, α2–β2, or α3–β2 isozymes or SERCA2a. After metabolic labeling and various pulse–chase periods, microsomes were prepared and subjected to immunoprecipitations under nondenaturing conditions with a NKA α- or SERCA-antibody (Fig. 1A) or directly loaded on gels to determine PLM expression levels (Fig. 1B). Under these latter conditions, a labeled band of about 13 kDa became apparent among oocyte proteins, which corresponded to PLM because it was absent in noninjected oocytes (data not shown) and could be immunoprecipitated with PLM-antibodies (see Fig. 1C). The α-antibody coimmunoprecipitated the 13-kDa PLM with all tested NKA isozymes after the pulse and a prolonged chase period (Fig. 1A, lanes 1–12), although somewhat less efficiently with α–β2 isozymes (lanes 7–12) than with α–β1 isozymes (lanes 1–8). On the contrary, PLM coexpressed with SERCA2a was not coimmunoprecipitated with a SERCA-antibody (Fig. 1A, lanes 13 and 14) despite a high expression of both SERCA2a (Fig. 1A, lanes 13 and 14) and PLM (Fig. 1B, lanes 13 and 14), indicating that association of PLM is specific for NKA isozymes.
Fig 1.
Association of PLM with NKA isozymes in Xenopus oocytes. (A) Oocytes were injected with 8 ng of rat NKA α1, α2, or α3 cRNA and 1 ng β1 or β2 cRNA, or with 8 ng SERCA2a cRNA together with 1 ng PLM cRNA, metabolically labeled for 24 h, and subjected to a 48-h chase period. After the pulse–chase period, microsomes were prepared and immunoprecipitated with an α- (lanes 1–12) or SERCA-antibody (lanes 13 and 14) under nondenaturing conditions before subjecting to SDS/PAGE. Indicated are the positions of PLM (≈13 kDa), NKA α isoforms, SERCA2a and fully (fg) and core glycosylated (cg) β isoforms. (B) Microsomes were directly loaded on a SDS/Tricine gel. (C) Oocytes were injected with NKA α1, β1, and PLM cRNA and metabolically labeled for 24 h before microsomes were prepared and immunoprecipitations were performed with an α- (lane 1) or a PLM-antibody (lane 2) under nondenaturing conditions. (D) After injection with NKA α1, β1, and PLM cRNA, oocytes were metabolically labeled in the absence (lane 1) or presence (lanes 4 and 7) of BFA and subjected to 24- and 48-h chase periods in the absence (lanes 2, 3, 5, and 6) or presence (lanes 8 and 9) of BFA. Microsomes were prepared and immunoprecipitated with an α-antibody under nondenaturing conditions. (E) Quantification of data shown in D. Represented is the ratio PLM/α as a function of the chase period; open circles, data from D, lanes 1–3; closed circles, data from D, lanes 4–6; closed triangles, data from D, lanes 7–9.
Interestingly, in oocytes coexpressing α1–β1 complexes and PLM, the PLM-antibody did coimmunoprecipitate the fully glycosylated but not the endoplasmic reticulum (ER)-located, core glycosylated β subunit (Fig. 1C, lane 2), which is still present after a 24-h pulse and is associated with the α subunit as revealed by immunoprecipitations with an α-antibody (lane 1). This result indicated that association of PLM with NKA occurs posttranslationally in the Golgi compartment or the plasma membrane. To test this hypothesis, we probed the association of PLM with NKA after a 6-h pulse, when the β subunit is mainly in its core glycosylated form, and after 24- and 48-h chase periods either in the absence or presence of brefeldin A (BFA). In Xenopus oocytes, BFA leads to redistribution into the ER of cis Golgi compartments but not of medial/trans Golgi compartments involved in complex type glycosylation, and to a block of protein transport from the ER out of the mixed ER/Golgi complex (27). Fig. 1D shows that, as expected, in the presence of BFA, the β subunit associated with the α subunit was exclusively present in its core glycosylated form (lane 4 and 7) after a 6-h pulse, whereas in the absence of BFA, part of the β subunit became fully glycosylated indicating its ER exit (lane 1). The effect of BFA was reversible because removal of BFA after the pulse period permitted full glycosylation of the β subunit (compare lanes 5 and 6 to lanes 8 and 9). Despite a similar expression of PLM after the pulse and the chase periods (data not shown), less PLM was coimmunoprecipitated with an α-antibody after a 6-h pulse (lanes 1, 4, and 7) than after a 24- (lanes 2, 5, and 8) or 48-h (3, 6, and 9) chase period, both in the absence and presence of BFA, supporting the idea of posttranslational association of PLM. By taking into account the number of leucine residues available for metabolic labeling in PLM and α subunits, quantifications of α and PLM indeed show that, in the absence of BFA, the PLM/α ratio is low after a 6-h pulse and only reaches a 1:1 stoichiometry after a 24-h chase (Fig. 1E). Moreover, in the presence of BFA, when all α–β complexes are retained in the ER, association is very inefficient but can again proceed when BFA is removed, which permits ER exit of α–β complexes (Fig. 1E). Similar results were obtained when α subunits were coimmunoprecipitated with a PLM-antibody (data not shown). These results show that, in Xenopus oocytes, PLM associates with NKA in a post-ER compartment in a 1:1 ratio and thus suggest that PLM has either different kinetics of ER exit or cell compartment-specific requirements for NKA association.
Association of PLM with NKA in Native Tissues.
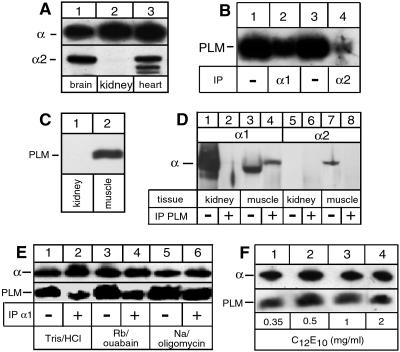
Fig. 2 documents our findings on the association of PLM with α isoforms present in membranes of bovine heart and rat skeletal muscle. As expected (5), the α1 isoform was detected in rat brain, rat kidney, bovine heart sarcolemma, and rat skeletal muscle, whereas the α2 isoform was found in brain, heart, and muscle but not in kidney (Fig. 2 A and D). There appears to be some degradation of the α2 isoform in the bovine heart membranes (Fig. 2A, lane 3). (In rat cardiac membranes the α2 isoform was completely degraded, precluding its use in coimmunoprecipitation experiments.) Fig. 2B shows that either an α1- or α2-specific antibody was able to coimmunoprecipitate PLM from the detergent-solubilized microsomes, although the efficiency appears to be higher for the anti-α1 compared with the anti-α2. The presence of PLM (Fig. 2C) and its association with NKA could also be demonstrated in microsomes of skeletal muscle (Fig. 2D). A PLM-antibody coimmunoprecipitated α1 isoforms from skeletal muscle (Fig. 2D, lane 4) but not from kidney microsomes (lane 2). Coimmunoprecipitation of α2 isoforms could not be revealed (lane 8), possibly because of the lower expression of α2 compared with α1 isoforms. The lower efficacy of coimmunoprecipitation of PLM and α2 isoforms in heart and skeletal muscle could reflect the predominant presence in these tissues of α2–β2 isozymes (5), which as shown in Fig. 1A associate less efficiently with PLM than α–β1 isozymes, or differences in the association efficiency between PLM and different NKA isozymes in the native tissue and Xenopus oocytes.
Fig 2.
Association of PLM with NKA isozymes in native tissues. (A) Aliquots of rat brain, rat kidney, or bovine sarcolemma microsomes were migrated on SDS/Tricine gels and subjected to Western blotting. Blots were probed with an α-antibody (anti-KETYY, Upper) or an α2-specific antibody (anti-HERED, Lower). (B) Aliquots of bovine sarcolemma microsomes were suspended in a solution containing 25 mM imidazole (pH 7.5), 1 mM EDTA, and 10 mM RbCl plus 5 mM ouabain and solubilized at 0°C with 1 mg/ml of C12E10. Samples were either loaded directly on SDS/Tricine gels (lanes 1 and 3) or first immunoprecipitated with an α1- (lane 2) or α2-specific (lane 4) antibody under nondenaturing conditions. After Western blotting, PLM was revealed with PLM-antibodies. (C) Aliquots of rat kidney or skeletal muscle microsomes were migrated on SDS/Tricine gels and subjected to Western blotting. Blots were probed with a PLM-antibody. (D) Aliquots of microsomes of kidney (lanes 1, 2, 5, and 6) or rat skeletal muscle (lanes 3, 4, 7, and 8) were either directly loaded on a gel (lanes 1, 3, 5, and 7) or first immunoprecipitated with a PLM-antibody under nondenaturing conditions (lanes 2, 4, 6, and 8). After Western blotting, α1 (lanes 1–4) and α2 isoforms (lanes 5–8) were revealed with specific antibodies. For unknown reasons, α1 isoforms from muscle (lane 3) migrated slightly faster than α1 from kidney (lane 1) or α1 coimmunoprecipitated with PLM-antibodies (lane 4). (E) Aliquots of sarcolemma microsomes were suspended in the presence of either 20 mM Tris⋅HCl, 10 mM RbCl plus 5 mM ouabain, or 20 mM NaCl plus 0.1 mg/ml of oligomycin before solubilization as in B. Samples were immunoprecipitated (lanes 2, 4, and 6) or not (lanes 1, 3, and 5) with a α1-specific antibody and subjected to Western blotting. Blots were probed with α- (Upper) or PLM-antibodies (Lower). (F) Aliquots of sarcolemma microsomes were suspended in the presence of 10 mM RbCl plus 5 mM ouabain and solubilized with the indicated concentrations of C12E10. Immunoprecipitation and Western blotting were as in E.
We have shown recently that the efficiency of coimmunoprecipitation of the γ subunit, and especially of CHIF, by the anti-α1 antibody is optimal when the membranes are solubilized with C12E10 in the presence of Rb+ plus ouabain or Na+ plus oligomycin. As emphasized in ref. 12, these conditions are required to preserve an intact structure of the α subunit, and the α/β/γ or α/β/CHIF complexes on detergent solubilization, and are not per se indicative of conformational dependence of the subunit interactions. The efficiency of coimmunoprecipitation of PLM was marginally better in a medium containing Na+/oligomycin compared with the Tris⋅HCl and Rb+/ouabain media (Fig. 2E). In addition, by contrast to the case of the γ subunit or CHIF, increasing concentrations of C12E10 did not abolish coimmunoprecipitation of PLM (Fig. 2F). The dependence of immunoprecipitation on the ionic conditions and detergent concentrations appears to reflect, in a qualitative way, the strength of the interaction of the FXYD protein with the α subunit. By either criterion, the order from the strongest to the weakest interaction is PLM > γ > CHIF.
Functional Effects of PLM on NKA Transport Properties.
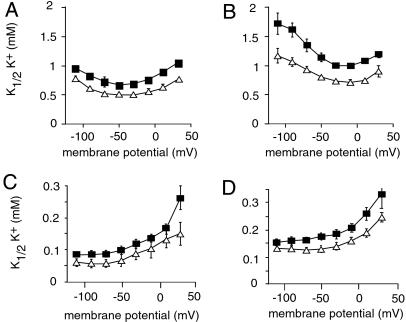
The functional effects of PLM on NKA were investigated by electrophysiological measurements of NKA transport properties in Xenopus oocytes expressing rat NKA α1–β1 or α2–β1 isozymes, which are present in heart and skeletal muscle (33, 34) together with PLM. As shown in Fig. 3 and Table 1, PLM produced a small but significant decrease in the apparent affinity for extracellular K+ of both NKA isozymes over a wide range of membrane potentials. The effect of PLM on the K+-activation kinetics measured in the absence or presence of external Na+ was slightly different for the two isozymes. PLM had a similar effect on the K+-activation kinetics with (Fig. 3A) and without (Fig. 3C) external Na+ over the whole potential range (Fig. 3E), suggesting a modification of the intrinsic affinity of the external K+ binding site. On the other hand, in PLM-associated α2–β1 complexes (Fig. 3 B and D), the apparent K+ affinity was decreased to a greater extent by the presence of external Na+ at high negative membrane potentials (Fig. 3F), suggesting that the translocation and the release of Na+ is also affected by the presence of PLM.
Fig 3.
PLM modulates the apparent K+ affinity of NKA isozymes. Oocytes were injected with PLM cRNA (1 ng) together with rat NKA α isoforms (α1, α2*, 10 ng) and rat β1 (1 ng) cRNAs. The voltage dependence of the external K+-activation constant (K1/2K+) of NKA α1–β1 (A and C) or α2–β1 (B and D) isozymes was measured in the presence (A and B) or absence (D and E) of 90 mM external Na+. PLM had a significant (P < 0.05) effect on the K+ activation of α1–β1 and α2–β1 isozymes over the whole potential range in the presence of external Na+. Open triangles, NKA alone; closed squares, NKA plus PLM. Data are means ± SE of 11–17 oocytes from two to three different batches.
Table 1.
Effect of PLM on transport properties (measured at −50 mV) and ouabain binding of NKA isoforms
| Isoform
|
K1/2K+, mM |
K1/2Na+, mM
|
Kd ouabain, nM
|
|
|---|---|---|---|---|
| +Na | −Na | |||
| α1–β1 | 0.49 ± 0.01 | 0.07 ± 0.02 | 9.3 ± 1.1 | 7.8 ± 0.5 |
| α1–β1–PLM | 0.67 ± 0.02 | 0.10 ± 0.01 | 16.5 ± 1.2 | 5.7 ± 1.3 |
| α2–β1 | 0.80 ± 0.03 | 0.14 ± 0.02 | 13.6 ± 1.6 | 38.4 ± 11 |
| α2–β1–PLM | 1.14 ± 0.07 | 0.18 ± 0.02 | 20.5 ± 2.5 | 50.3 ± 15 |
, P < 0.05 compared to PLM-less NKA isozyme.
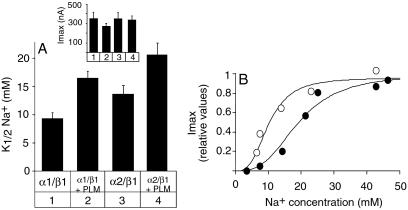
PLM association had an even more pronounced effect on the apparent affinity for internal Na+ of both α1–β1 and α2–β1 isozymes (Fig. 4 and Table 1). Association of PLM induced a 1.5- to 2-fold decrease in the apparent Na+ affinity of α1–β1 (Fig. 4A, lanes 1 and 2) and α2–β1 (lanes 3 and 4) isozymes, whereas maximal Na,K-pump currents were not significantly affected (Fig. 4A Inset). Significantly, as reflected by the Na+-activation curve (Fig. 4B), at high intracellular Na+ concentrations (e.g., 20 mM), NKA associated with PLM has an important spare transport capacity, whereas NKA lacking PLM transports at a nearly maximal rate.
Fig 4.
PLM decreases the apparent Na+ affinity of NKA isozymes. Oocytes were injected with rat NKA α1 and β1 (lanes 1 and 2) or α2* and β1 (lanes 3 and 4) cRNAs without (lanes 1 and 3) or with PLM (lanes 2 and 6) cRNA in the presence of cRNAs of the rat epithelial Na+ channel subunits (α, β, γ, 0.3 ng each), and incubated for 3 days. Apparent affinities for intracellular Na+ shown in A were determined as described in Materials and Methods from Na+-activation curves shown in B. Open circles, NKA α1–β1; closed circles, NKA α1–β1 plus PLM. PLM had a significant effect (P < 0.05) on the apparent Na+ affinity of each NKA isozyme tested. (Inset) Maximal pump currents (Imax). Data are means ± SE of 5–11 oocytes from two to three different batches.
Because NKA in the heart is the target for cardiac glycosides and possibly endoouabain, we finally investigated the effect of PLM association on the ouabain affinity of α1–β1 and α2–β1 isozymes. As shown in Table 1, the Kd value for ouabain of the two NKA isozymes was similar when expressed in the presence or absence of PLM.
Discussion
In the present paper, we provide evidence that PLM may be a tissue-specific regulator of NKA similar to other members of the FXYD protein family such as the γ subunit of NKA (14, 35, 36), CHIF (12), and FXYD7 (44). Indeed, our results show that PLM is associated with NKA in heart and skeletal muscles and that it modifies the transport properties of different NKA isozymes after coexpression in Xenopus oocytes.
As other members of the FXYD family, the FXYD1 protein, PLM, is a type I membrane protein (20) that exposes its C terminus to the cytoplasmic side. Similar to CHIF (13), but in contrast to the γ subunit (14) and FXYD7 (44), PLM adopts this membrane orientation after cleavage of a signal sequence (20). Despite intense investigation, the real physiological role of PLM has so far not been identified. PLM can be phosphorylated by several hormones—e.g., by insulin in skeletal muscle, by α- and β-adrenergic receptors in myocardial sarcolemma, and by vasopression in smooth muscles (for references see ref. 37). Ser-63 and -68 in the cytoplasmic C terminus were shown to be differential targets for PKA, PKC, and NIMA kinase phosphorylation (for references see ref. 37) that probably play a distinct regulatory role in the function of PLM (37). Significantly, PLM shares with other FXYD proteins the ability to induce an ion conductance when expressed in Xenopus oocytes (38). Similar to Mat-8 (17), PLM induces hyperpolarization-activated Cl-selective currents (38). Moreover, both when expressed in oocytes or added to lipid bilayers, PLM selectively transports the zwitterionic amino acid taurine (22), an osmolyte of animal cells. Recently, it was also shown that PLM stably transfected in HEK cells increases taurine efflux and regulatory volume decrease in response to cell swelling (39) and that in astrocytes taurine efflux can be inhibited after a decrease in the expression of endogenous PLM by antisense oligonucleotide (40). Thus, based on its function as a channel or a channel regulator, it is believed that PLM plays a specific role in muscle contractility and cell volume regulation.
Our observation that PLM and other FXYD proteins interact with and modulate NKA adds a new perspective to the functional role of this protein family. In the case of PLM, expressed in skeletal and heart muscle, we can predict that the observed modulatory effect of PLM on the apparent Na+ affinity of NKA may be as favorable for efficient muscle contractility as the formation or regulation of ion channels. The existence of low Na+-affinity Na,K-pumps may indeed be necessary for an efficient extrusion of increased intracellular Na+ ([Na+]i) during action potentials in contractile tissues to permit appropriate excitability. Upon a rise in [Na+]i—e.g., up to 20 mM—NKA with a K1/2Na+ of about 10 mM would transport at a nearly maximal rate, whereas NKA associated with PLM with a K1/2Na+ of about 20 mM would only transport at half-maximal rates and would still have the capacity to increase its rate at higher [Na+]i. Significantly, in cardiac cells, K1/2Na+ for Na,K-pumps vary widely according to the experimental procedure and ionic conditions used (for review, see ref. 41). However, the K1/2Na+ of about 20 mM, which we determined for NKA associated with PLM, is close to that predicted for cardiac Na,K-pumps under physiological conditions (41). This provides an additional argument for the association of PLM with NKA in cardiac tissues and its physiological relevance. There is indeed increasing evidence that the variability of Na+ affinities of Na,K-pumps in different tissues is not only due to expression of different NKA isozymes, but also due to the association of tissue-specific FXYD proteins with NKA. For instance, in the kidney, Na+ affinities of NKA range between 3 and 10 mM in different nephron segments despite the sole presence of the α1–β1 isozyme (42). The different Na+ affinities of the renal α1–β1 isozyme along the nephron indeed correlate well with the distribution of CHIF and γ subunits and with the effects of these FXYD proteins on the apparent Na+ affinity of the α1–β1 isozyme (12, 13).
A challenging question that remains to be addressed concerns the factors that regulate the potentially dual function of PLM. So far, little is known on determinants that influence the expression of PLM or its channel or channel-regulating function. Coexpression of protein kinases with PLM in Xenopus oocytes indicates that phosphorylation of PLM by PKA, but not by PKC, largely increases the amplitude of oocyte currents as well as the amount of PLM in oocyte membranes, whereas similar effects induced by NIMA kinase do not require PLM phosphorylation (38). It will be interesting to compare the role played by phosphorylation and by hormonal stimulation in the regulatory effect of PLM on NKA. In this context, it is interesting to note that in cardiac myocytes, aldosterone decreases Na,K-pump currents independent of a change in total cellular or cell surface-expressed NKA levels (43). These results indicate that hyperaldosteronemia decreases the apparent Na+ affinity of cardiac NKA, an effect that mimics the effect of association of PLM with NKA. Perhaps this example reflects the possibility that in particular physiological or pathophysiological situations, specific hormonal control may favor either the NKA regulatory function or the ion channel-related function of PLM, or in general of FXYD proteins.
In conclusion, in this study we have identified another protein of the FXYD family, PLM, as a regulator of NKA. These results provide further evidence for a unique mode of regulation of NKA by FXYD proteins that implicates a tissue- and physiological state-specific expression of an auxiliary subunit of NKA.
Acknowledgments
We thank Danièle Schaer and Sophie Roy for excellent technical assistance. We also thank M. J. Caplan for the α1-antibody (6H), T. Pressley for the α1 and α2 isoform-specific antibodies, B. Attali for dog PLM cDNA, J. Lingrel for rat NKA α1, α2*, and α3* and β1 cDNAs, D. H. MacLennan for SERCA2a cDNAs, and D. Khananshvili for the bovine heart sarcolemma vesicles. This work was supported by grants from the Swiss National Fund for Scientific Research (31-64793.01 to K.G.) and the Roche Research Foundation (to K.G. and G.C.), and by a grant from the Minerva Foundation (to S.K. and H.G.).
Abbreviations
NKA, Na,K-ATPase
PLM, phospholemman
ER, endoplasmic reticulum
BFA, brefeldin A
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Gheorghiade M. & Pitt, B. (1997) Am. Heart J. 134, 3-12. [DOI] [PubMed] [Google Scholar]
- 2.Hamlyn J. M., Hamilton, B. P. & Manunta, P. (1996) J. Hypertens. 14, 151-167. [DOI] [PubMed] [Google Scholar]
- 3.Geering K. (2001) J. Bioenerg. Biomembr. 33, 425-438. [DOI] [PubMed] [Google Scholar]
- 4.Crambert G., Hasler, U., Beggah, A. T., Yu, C., Modyanov, N. N., Horisberger, J. D., Lelièvre, L. & Geering, K. (2000) J. Biol. Chem. 275, 1976-1986. [DOI] [PubMed] [Google Scholar]
- 5.Blanco G. & Mercer, R. W. (1998) Am. J. Physiol. 275, F633-F650. [DOI] [PubMed] [Google Scholar]
- 6.Feraille E. & Doucet, A. (2001) Physiol. Rev. 81, 345-418. [DOI] [PubMed] [Google Scholar]
- 7.Sweadner K. J. & Rael, E. (2000) Genomics 68, 41-56. [DOI] [PubMed] [Google Scholar]
- 8.Forbush B., Kaplan, J. H. & Hoffman, J. F. (1978) Biochemistry 17, 3667-3676. [DOI] [PubMed] [Google Scholar]
- 9.Mercer R. W., Biemesderfer, D., Bliss, D. P., Collins, J. H. & Forbush, B. (1993) J. Cell Biol. 121, 579-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuster B., Shainskaya, A., Pu, H. X., Goldshleger, R., Blostein, R., Mann, M. & Karlish, S. J. (2000) J. Biol. Chem. 275, 18441-18446. [DOI] [PubMed] [Google Scholar]
- 11.Attali B., Latter, H., Rachamim, N. & Garty, H. (1995) Proc. Natl. Acad. Sci. USA 92, 6092-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garty, H., Lindzen, M., Scanfano, R., Aizman, R., Füzesi, M., Goldshleger, R., Farman, N., Blostein, R. & Karlish, S. J. D. (2002) Am. J. Physiol., in press. [DOI] [PubMed]
- 13.Béguin P., Crambert, G., Guennoun, S., Garty, H., Horisberger, J.-D. & Geering, K. (2001) EMBO J. 20, 3993-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Béguin P., Wang, X. Y., Firsov, D., Puoti, A., Claeys, D., Horisberger, J. D. & Geering, K. (1997) EMBO J. 16, 4250-4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arystarkhova E., Donnet, C., Asinovski, N. K. & Sweadner, K. J. (2002) J. Biol. Chem. 277, 10162-10172. [DOI] [PubMed] [Google Scholar]
- 16.Pu H. X., Cluzeaud, F., Goldshlegger, R., Karlish, S. J. D., Farman, N. & Blostein, R. (2001) J. Biol. Chem. 276, 20370-20378. [DOI] [PubMed] [Google Scholar]
- 17.Morrison B. W., Moorman, J. R., Kowdley, G. C., Kobayashi, Y. M., Jones, L. R. & Leder, P. (1995) J. Biol. Chem. 270, 2176-2182. [DOI] [PubMed] [Google Scholar]
- 18.Fu X. & Kamps, M. (1997) Mol. Cell. Biol. 17, 1503-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamaguchi F., Yamaguchi, K., Tai, Y., Sugimoto, K. & Tokuda, M. (2001) Brain Res. Mol. Brain Res. 86, 189-192. [DOI] [PubMed] [Google Scholar]
- 20.Palmer C. J., Scott, B. T. & Jones, L. R. (1991) J. Biol. Chem. 266, 11126-11130. [PubMed] [Google Scholar]
- 21.Chen L. S. K., Lo, C. F., Numann, R. & Cuddy, M. (1997) Genomics 41, 435-443. [DOI] [PubMed] [Google Scholar]
- 22.Moorman J. R., Ackerman, S. J., Kowdley, G. C., Griffin, M. P., Mounsey, J. P., Chen, Z. H., Cala, S. E., Obrian, J. J., Szabo, G. & Jones, L. R. (1995) Nature (London) 377, 737-740. [DOI] [PubMed] [Google Scholar]
- 23.Kowdley G. C., Ackerman, S. J., Chen, Z. H., Szabo, G., Jones, L. R. & Moorman, J. R. (1997) Biophys. J. 72, 141-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahmmoud Y. A., Vorum, H. & Cornelius, F. (2000) J. Biol. Chem. 275, 35969-35977. [DOI] [PubMed] [Google Scholar]
- 25.Jewell E. A. & Lingrel, J. B. (1991) J. Biol. Chem. 266, 16925-16930. [PubMed] [Google Scholar]
- 26.Melton D. A., Krieg, P. A., Rebagliati, M. R., Maniatis, T., Zinn, K. & Green, M. R. (1984) Nucleic Acids Res. 12, 7035-7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geering K., Beggah, A., Good, P., Girardet, S., Roy, S., Schaer, D. & Jaunin, P. (1996) J. Cell Biol. 133, 1193-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones L. R. (1988) Methods Enzymol. 157, 85-91. [DOI] [PubMed] [Google Scholar]
- 29.Crambert, G., Béguin, P., Pestov, N. B., Modyanov, N. N. & Geering, K. (2002) Biochemistry, in press. [DOI] [PubMed]
- 30.Pressley T. A. (1992) Am. J. Physiol. 262, C743-C751. [DOI] [PubMed] [Google Scholar]
- 31.Jaisser F., Jaunin, P., Geering, K., Rossier, B. C. & Horisberger, J. D. (1994) J. Gen. Physiol. 103, 605-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hasler U., Wang, X., Crambert, G., Béguin, P., Jaisser, F., Horisberger, J. D. & Geering, K. (1998) J. Biol. Chem. 273, 30826-30835. [DOI] [PubMed] [Google Scholar]
- 33.Wang J. N., Schwinger, R. H. G., Frank, K., Mullerehmsen, J., Martin-Vasallo, P., Pressley, T. A., Xiang, A., Erdmann, E. & McDonough, A. A. (1996) J. Clin. Invest. 98, 1650-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson C. & McDonough, A. (1996) J. Biol. Chem. 271, 32653-32658. [DOI] [PubMed] [Google Scholar]
- 35.Therien A. G., Goldshleger, R., Karlish, S. J. & Blostein, R. (1997) J. Biol. Chem. 272, 32628-32634. [DOI] [PubMed] [Google Scholar]
- 36.Arystarkhova E., Wetzel, R. K., Asinovski, N. K. & Sweadner, K. J. (1999) J. Biol. Chem. 274, 33183-33185. [DOI] [PubMed] [Google Scholar]
- 37.Walaas S. I., Czernik, A. J., Olstad, O. K., Sletten, K. & Walaas, O. (1994) Biochem. J. 304, 635-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moorman J. R., Palmer, C. J., John, J. E., III, Durieux, M. E. & Jones, L. R. (1992) J. Biol. Chem. 267, 14551-14554. [PubMed] [Google Scholar]
- 39.Morales-Mulia M., Pasantes-Morales, H. & Moran, J. (2000) Biochim. Biophys. Acta 1496, 252-260. [DOI] [PubMed] [Google Scholar]
- 40.Moran J., Morales-Mulia, M. & Pasantes-Morales, H. (2001) Biochim. Biophys. Acta 1538, 313-320. [DOI] [PubMed] [Google Scholar]
- 41.Glitsch H. G. (2001) Physiol. Rev. 81, 1791-1826. [DOI] [PubMed] [Google Scholar]
- 42.Barlet-Bas C., Cheval, L., Khadouri, C., Marsy, S. & Doucet, A. (1990) Am. J. Physiol. 259, F246-F250. [DOI] [PubMed] [Google Scholar]
- 43.Mihailidou A. S., Bundgaard, H., Mardini, M., Hansen, P. S., Kjeldsen, K. & Rasmussen, H. H. (2000) Circ. Res. 86, 37-42. [DOI] [PubMed] [Google Scholar]
- 44.Béguin P., Crambert, G., Monnet-Tschudi, F., Uldry, M., Horisberger, J.-D., Garty, H. & Gerring, K. (2002) EMBO J. 21, 3264-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]