Abstract
Stearoyl–CoA desaturase (SCD) is a central lipogenic enzyme catalyzing the synthesis of monounsaturated fatty acids, mainly oleate (C18:1) and palmitoleate (C16:1), which are components of membrane phospholipids, triglycerides, wax esters, and cholesterol esters. Several SCD isoforms (SCD1-3) exist in the mouse. Here we show that mice with a targeted disruption of the SCD1 isoform have reduced body adiposity, increased insulin sensitivity, and are resistant to diet-induced weight gain. The protection from obesity involves increased energy expenditure and increased oxygen consumption. Compared with the wild-type mice the SCD1−/− mice have increased levels of plasma ketone bodies but reduced levels of plasma insulin and leptin. In the SCD1−/− mice, the expression of several genes of lipid oxidation are up-regulated, whereas lipid synthesis genes are down-regulated. These observations suggest that a consequence of SCD1 deficiency is an activation of lipid oxidation in addition to reduced triglyceride synthesis and storage.
Stearoyl–CoA desaturase (SCD) is the rate-limiting enzyme in the biosynthesis of monounsaturated fatty acids. It catalyzes the introduction of the cis double bond in the Δ9 position of fatty acyl–CoA substrates. The preferred desaturation substrates are palmitoyl–CoA and stearoyl–CoA, which are converted to palmitoleoyl–CoA (16:1) and oleoyl–CoA (18:1), respectively (1–4). These fatty acids are requisite components of membrane phospholipids, triglycerides, cholesterol esters, and wax esters (5–7). Effects on composition of phospholipids ultimately determine membrane fluidity, and the effects on the composition of cholesterol esters and triglycerides can affect lipoprotein metabolism and adiposity. SCD expression is sensitive to dietary factors including polyunsaturated fatty acids, cholesterol and vitamin A, hormonal changes (i.e., insulin and glucagon), developmental processes, temperature changes, thiazolinediones, metals, alcohol, peroxisomal proliferators, and phenolic compounds (3). High SCD activity has been implicated in a wide range of disorders including diabetes, atherosclerosis, cancer, obesity, and viral infection (3, 8–13).
The existence of multiple SCD isoforms in mice (6, 14–18) and rats makes it difficult to determine the role of each isoform in lipid metabolism. New insights into the physiological role of the SCD1 gene and its endogenous products came from recent studies of the asebia mouse strains (abj and ab2j) that have naturally occurring mutations in SCD1 (17–19) as well as a laboratory mouse model with a targeted disruption (SCD1−/−) (6). We used these animal models to show that SCD1−/− mice are deficient in hepatic triglycerides and cholesterol esters (7, 20). The levels of palmitoleate (16:1) and oleate (18:1) are reduced, whereas palmitate and stearate are increased in the lipid fractions of SCD1−/− mice. On a high carbohydrate diet supplemented with triolein, the cholesterol ester levels are corrected but the triglyceride levels are not reversed to the levels found in the wild-type mouse (7).
Apart from the dramatic alterations in triglyceride and cholesterol metabolism, the SCD1−/− mice are considerably leaner than their wild-type counterparts. Here, we show changes in metabolic rate and in the expression of genes encoding enzymes involved in lipid metabolism.
Methods
Animals and Diets.
SCD1−/− mice in SV129 background were generated and genotyped as described (5). The wild-type (SCD1+/+), heterozygous (SCD1+/−) and homozygous (SCD1−/−) mice are housed and bred in a pathogen-free barrier facility of the Department of Biochemistry (Univ. of Wisconsin, Madison) operating at room temperature in a 12-h light/12-h dark cycle. The breeding of these animals was in accordance with the protocols approved by the animal care research committee of the Univ. of Wisconsin. At 3 weeks of age, the mice were fed ad libitum a standard laboratory chow diet or a high-fat diet for 23 weeks. The high-fat diet contains 195 g/kg casein, 3 g/kg DL-methionine, 377 g/kg sucrose, 150 g/kg corn starch, 153 g/kg anhydrous milkfat, 10 g/kg corn oil, 1.5 g/kg cholesterol, 60.067 g/kg cellulose, 35 g/kg mineral mix AIN-76 (170915), 4 g/kg calcium carbonate, 10 g/kg vitamin mix Teklad (40060), 1.2 g/kg choline bitartrate, and 0.033 g/kg ethoxyquin (antioxidant).The weight of each mouse within each group was measured weekly; the data are presented as means ± SD (n = 8, P < 0.001). The glucose tolerance and insulin tolerance were determined as described (21).
Measurement of Oxygen Consumption.
Gender matched SCD1−/− and wild-type littermates were investigated in indirect calorimeters as described (22). Oxygen consumption rate (VO2) and CO2 production rate (VCO2) were continuously assayed over 4 consecutive 23-h periods, including 12 h dark (1800–0600) and 11 h light (0600–1700).
Gene Expression Analysis.
RNA was isolated from livers of 10 individual 6-week-old female mice by using a standard method (23). Mouse genome U74A arrays were used to monitor the expression level of approximately 12,000 genes and expressed sequence tags (Affymetrix). Genes differentially expressed were identified by comparing expression levels in SCD1−/− and wild-type mice (24, 25). For Northern blot analysis, 20 μg of total liver RNA was separated on an 0.8% agarose/formaldehyde gel, transferred onto nylon membrane, and hybridized with cDNA probes for the corresponding genes.
Results
Reduced Body Weight in SCD1−/− Mice Fed a High-Fat Diet.
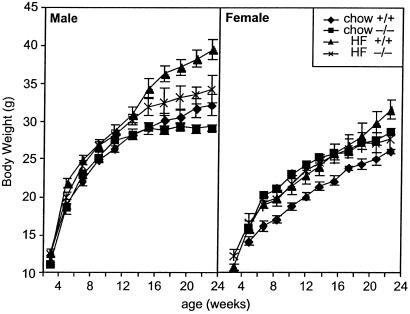
Although the growth curves of male SCD1−/− mice were similar to those of wild-type siblings on chow diet, a high-fat diet revealed large differences in weight gain in both males (34.2 g vs. 39.5 g, P < 0.01, Fig. 1) and females (27.7 g vs. 31.9 g, P < 0.05).
Fig 1.
Body weight of male and female wild-type and SCD1−/− mice fed a chow or high-fat diet.
Reduced Body Fat Mass in SCD1−/− Mice.
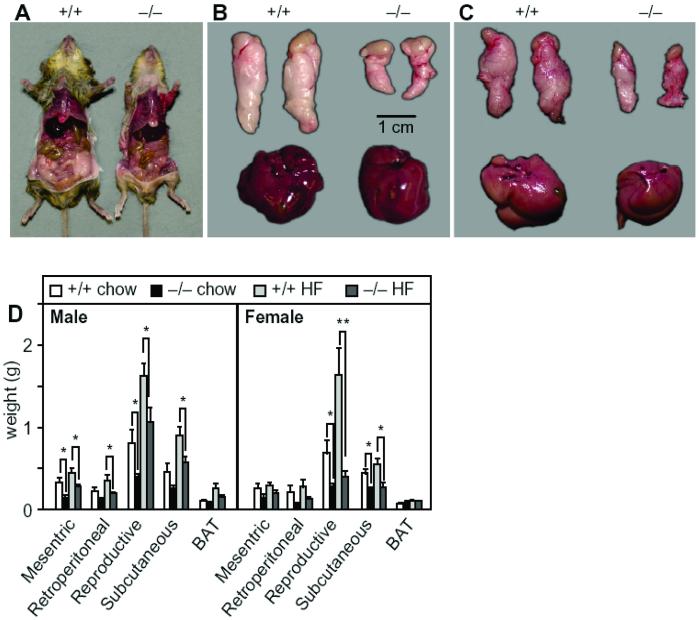
On average, the SCD1−/− mice consumed 25% more food than wild-type mice (4.1 g/day vs. 5.6 g/day; n = 9, P < 0.05). Nonetheless, they were leaner and accumulated less fat in their adipose tissue (Fig. 2A). The epididymal fat pad mass was markedly reduced in male SCD1−/− relative to wild-type mice fed a chow diet (0.4 ± 0.1 mg vs. 0.8 ± 0.2; n = 9, P < 0.05; Fig. 2B) and a high-fat diet (1.0 ± 0.2 mg vs. 1.6 ± 0.2, n = 12, P < 0.05; Fig. 2C). The livers of the wild-type and SCD1−/− mice were grossly normal and of similar mass. In contrast, on a high-fat diet, the livers of the wild-type mice were lighter in color than those of the mutant mice (Fig. 2C), suggestive of hepatic steatosis. The masses of white adipose depots in SCD1−/− mice were globally reduced in mice on either the chow or the high-fat diet (Fig. 2D). The masses of other tissues, including brown adipose tissue, were not significantly altered. Thus, SCD1−/− mice were resistant to diet-induced weight gain and fat accumulation, despite increased food intake.
Fig 2.
(A) Abdominal view of the fat pad under the skin in 23-week-old male wild-type and SCD1−/− mice. (B) Epididymal fat pads and liver isolated from the wild-type and SCD1−/− mice on a chow diet. (C) Epididymal fat pads and liver isolated from the wild-type and SCD1−/− mice on a high-fat diet. (Bar = 1 cm.) (D) Fat pad weights from mice fed chow and high-fat diets.
Increased Oxygen Consumption in SCD1−/− Mice.
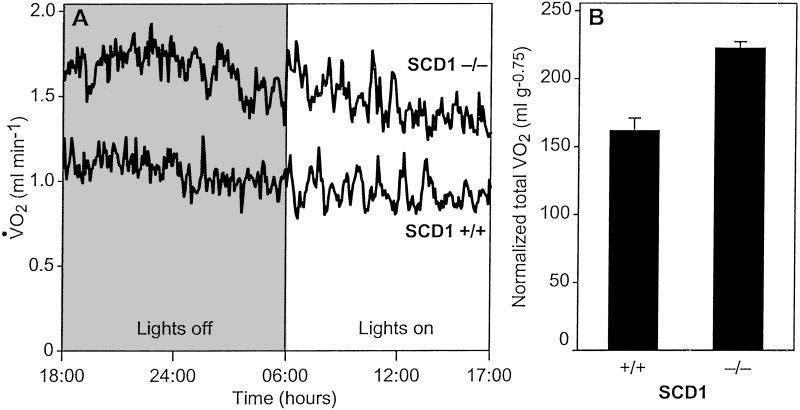
We carried out indirect calorimetry to investigate whether the resistance to weight gain is caused by increased energy expenditure. The SCD1−/− mice exhibited consistently higher rates of oxygen consumption (had higher metabolic rates) than their wild-type littermates throughout the day and night (Fig. 3A). After adjusting for allometric scaling and gender, the effect of the knockout allele was highly significant (P = 0.00019, multiple ANOVA, Fig. 3B).
Fig 3.
(A) Metabolic rate and oxygen consumption of male mice on a chow diet. (B) Gender-adjusted, normalized total oxygen consumption over a 23-h period. Error bars denote SE.
Because the increase in O2 consumption occurred during the fasting phase (daytime) as well as during the feeding phase, the animals are more active in oxidizing fat. Although ketone bodies were undetectable in plasma from either strain during postprandial conditions, β-hydroxybutyrate levels were much higher in the SCD1−/− mice after a 4-h fast (4.4 ± 0.6 mg/dl vs. 1.1 ± 0.7 mg/dl; P < 0.001), indicating a higher rate of β-oxidation in knockout mice. A similar but less dramatic difference was seen in females. These differences were also observed in mice on high-fat diet.
Increased Expression of Genes Involved in Fatty Acid Oxidation in SCD1−/− Mice.
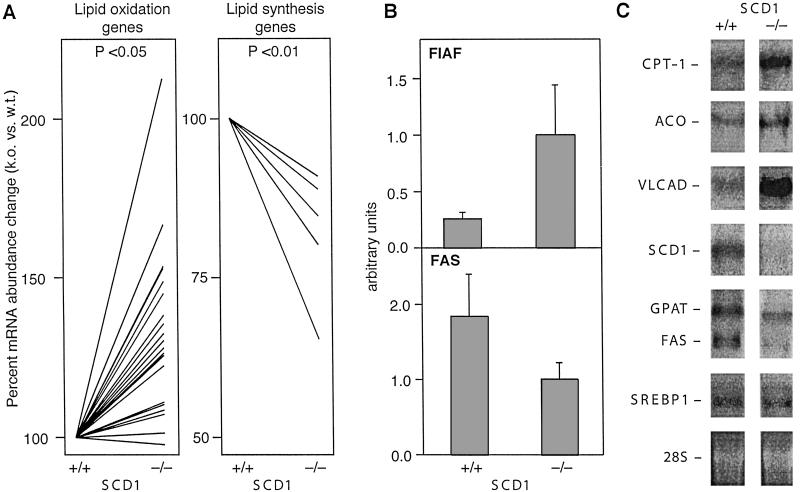
We used DNA microarrays to identify genes whose expression was altered in the livers of SCD1−/− mice. We identified 200 mRNAs that were significantly different between the livers of SCD1−/− and wild-type mice. The most striking pattern was seen in genes involved in lipogenesis and fatty acid β-oxidation. Lipid oxidation genes were up-regulated, whereas lipid synthesis genes were down-regulated in the SCD1−/− mice (Fig. 4A). Using the same RNA samples, the microarray data were verified with quantitative reverse-transcription–PCR using DNA primers that were designed for selected genes that showed differential expression (26). The results showed that the PPARα-target gene Fasting-Induced Adipocyte Factor (FIAF) was up-regulated in SCD1−/− mice (P < 0.05; Fig. 4B), whereas fatty acid synthase (FAS) was down-regulated (P < 0.01).
Fig 4.
(A) Expression levels of lipid oxidation (Left) and lipid synthesis (Right) genes between wild-type and SCD1−/− mice. (B) Quantitative reverse-transcription–PCR of FIAF and FAS gene expression, relative to wild-type mice. 18S RNA was used as a normalization control. (C) Northern blot analysis of lipid oxidation genes and lipid synthesis genes (SREBP-1, FAS, and GPAT) in the wild-type and SCD1−/− mice.
Northern blot analysis also supports changes in fatty acid oxidation and lipid biosynthesis. Probes for acyl–CoA oxidase (ACO), very long chain acyl–CoA dehydrogenase (VLCAD), and carnitine palmitoyltransferase-1 (CPT-1) indicate increases in β-oxidation (27, 28), whereas probes for SREBP-1, FAS, and glycerol phosphate acyl–CoA transferase (GPAT) point to a decrease in triglyceride biosynthesis (Fig. 4C).
Increased Insulin Sensitivity in SCD1−/− Mice.
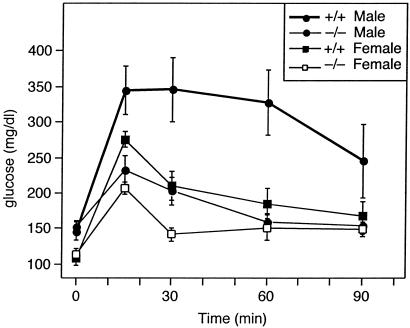
Reduced adipose tissue mass could either elicit insulin resistance or insulin sensitivity as demonstrated in several animal models (28). Fasting insulin levels were lower in the male SCD1−/− on chow diet (1.3 ± 0.3 ng/dl; n = 7) compared with wild-type mice (2.5 ± 0.9 ng/ml; n = 7). On a high-fat diet, insulin levels were similar between the two groups. Fasting glucose levels were similar between the SCD1−/− and wild-type mice. However, male and female SCD1−/− mice showed improved glucose tolerance compared with wild type (Fig. 5, P < 0.05). Thirty minutes after a glucose load, both male and female SCD1−/− mice tended to have lower fasting glucose levels (males: wild type, 345 ± 44 mg/dl; SCD1−/− mice, 202 ± 20, n = 8; females: wild type, 209 ± 20; SCD1−/− mice, 141 ± 9, n = 5). In addition, the glucose lowering effect of insulin was greater in the SCD1−/− mice than wild-type mice (data not shown). These data indicate that SCD1−/− mice have increased insulin sensitivity along with their loss of adiposity.
Fig 5.
Plasma glucose levels during the glucose tolerance test of male and female wild-type and SCD1−/− mice.
Discussion
These studies establish a critical role for SCD in the generation of body fat. The deletion of the SCD1 gene resulted in global changes in gene expression and altered metabolic activity that can account for the loss of body fat.
Genes encoding enzymes that participate in fatty acid oxidation were up-regulated in the SCD1−/−mice. CPT-1, ACO, VLCAD, and FIAF are known targets of PPARα (27, 28) and contain PPARα response regions in their promoters (28). Because PPARα mRNA is unchanged (data not shown), the up-regulation of enzymes of fatty acid β-oxidation in the SCD1−/− mice must be downstream of PPARα transcription. Thus, it is possible that loss of SCD1 function results in an increase in the concentration of a PPARα activator, perhaps a lipid ligand. The contents of saturated fatty acids (C16:0 and C18:0) are increased, whereas the contents of the polyunsaturated fatty acids of the n-6 and n-3 are not changed in the liver of the SCD1−/− mice (8, 10). One possible mechanism for our observations is that the saturated fatty acids induce the signal that activates the PPARα in the SCD1−/− mice, but this has yet to be determined. Alternatively the increased levels of C18:0- or C16:0-CoAs could inhibit acetyl–CoA carboxylase (ACC) through a well-known feedback mechanism; the resulting drop in malonyl–CoA can derepress CPT-1, resulting in increased transport of fatty acids into the mitochondria. Thus, the mechanism of increased lipid oxidation in the SCD1-deficient mouse could be caused by induction of PPARα-target genes as well enhanced availability of fatty acids for mitochondrial β-oxidation.
The SCD1−/− mice showed decreased expression in the liver of lipogenic genes SREBP-1, FAS, and GPAT (Fig. 4C). SREBP-1c is the main SREBP-1 isoform expressed in the liver and regulates the expression of lipogenic genes (29). Insulin, dietary carbohydrate, fatty acids, and cholesterol regulate SREBP-1 gene expression and protein maturation (29, 30). Thus, the down-regulation of SREBP-1 gene expression in the SCD1−/− mice could have numerous effects on various metabolic pathways regulated by SREBP-1. For instance the induction of SREBP-1 by insulin and cholesterol greatly enhances the synthesis and secretion of triglycerides by the liver (31). However, in the SCD1 knockout mice, carbohydrate feeding fails to induce triglyceride synthesis and secretion by the liver (7, 20). In addition, the SCD1 deficiency attenuates triglyceride synthesis and very low density lipoprotein secretion in the ob/ob mouse (32), implying that SCD1 represents a crucial bottleneck in triglyceride synthesis in the mouse.
In contrast to human subjects and several mouse models of lipodystrophy (33–36), the loss of adiposity in the SCD1−/− mice led to increased rather than decreased insulin sensitivity. In lipodystrophy, there is a redistribution in the lipogenic burden away from adipose tissue, leading to triglyceride accumulation in the liver and in skeletal muscle. Skeletal muscle triglyceride levels have recently been shown to strongly correlate with impaired insulin-stimulated glucose disposal. The reduction in muscle triglyceride content (M. Rahman, M.M., and J.M.N., unpublished data) in the SCD1−/− mice may contribute to increased insulin sensitivity observed in these mice.
Lipodystrophic Crebbp heterozygous null mice (37) have increased energy expenditure and unlike other lipodystrophic mouse models, increased insulin sensitivity. This has been attributed to increased plasma leptin levels. We measured plasma leptin to determine whether changes in levels of plasma leptin could account for the protection from weight gain, increased energy expenditure and insulin sensitivity in the SCD1−/− mice. Plasma leptin was significantly reduced in the SCD1−/− mice relative to the wild-type controls (on chow diet: males, 5.0 ± 0.5 vs. 25.3 ± 5.5 ng/ml, P < 0.01; females, 5.1 ± 0.9 ng/ml vs. 11.1 ± 1.2 ng/ml, P < 0.001). A similar large difference was observed in mice on high-fat diet. Plasma leptin remained lower in SCD1−/− mice even after correcting for reduced fat mass. Thus, the SCD1−/− mouse does not resemble the Crebbp+/− mouse, because the protection from adiposity is present despite lower leptin levels. These data suggest that SCD1 acts downstream of leptin, and predict that loss of SCD function would ameliorate the severe obesity observed in leptin-deficient ob/ob mice. Indeed, double mutant asebia ob/ob mice weighed significantly less than C57BL/6–ob/ob mice (32).
In conclusion, our studies have revealed that SCD1 gene deficiency leads to resistance to diet-induced obesity, increased insulin sensitivity, and increased metabolic rate. Because leptin represses the expression of the SCD1 gene and the SCD1 deficiency normalizes the hypometabolic phenotype of the ob/ob mice (32), our results are consistent with SCD1 being a target of leptin signaling, as suggested by the gene array studies of Soukas et al. (38) and confirmed by Cohen et al. (32). In addition, the expression of PPARα target genes of lipid oxidation were up-regulated in mouse liver of SCD1−/− mice, whereas those of SREBP-1 target genes of lipid synthesis were down-regulated. The studies suggest that SCD1 deficiency either directly or indirectly induces a signal that activates the PPARα pathway to partition fat toward oxidation and down-regulates SREBP-1 expression thereby reducing lipid synthesis and storage. These metabolic changes recommend SCD as a promising therapeutic target for the many disorders associated with the metabolic syndrome.
Acknowledgments
We thank Dr. Jeffrey Peters for the cDNAs for ACO, VLCAD, CPT-1, and PPARα. We thank Mary Rabaglia for assistance with insulin measurements. We thank Yeonhwa Park for critical reading of this manuscript. This work was supported in part by National Institutes of Health Medical Scientist Training Program Grant GM07739 (to P.C.), National Institutes of Health Grants R01DK-41096 (to J.M.F.), HL-56593 and R01DK-58037 (to A.D.A.), and R01DK-62388 (to J.M.N.), and by Xenon Genetics, Inc. (to M.M.).
Abbreviations
SCD, stearoyl–CoA desaturase
References
- 1.Ntambi J. M. (1999) J. Lipid Res. 40, 1549-1558. [PubMed] [Google Scholar]
- 2.Enoch H. G., Catala, A. & Strittmatter, P. (1976) J. Biol. Chem. 251, 5095-5103. [PubMed] [Google Scholar]
- 3.Ntambi J. M. (1995) Prog. Lipid Res. 34, 139-150. [DOI] [PubMed] [Google Scholar]
- 4.Kasturi R. & Joshi, V. C. (1982) J. Biol. Chem. 257, 12224-12230. [PubMed] [Google Scholar]
- 5.Miyazaki M., Man, W. C. & Ntambi, J. M. (2001) J. Nutr. 131, 2260-2268. [DOI] [PubMed] [Google Scholar]
- 6.Miyazaki M., Kim, H. J., Man, W. C. & Ntambi, J. M. (2001) J. Biol. Chem. 276, 39455-39461. [DOI] [PubMed] [Google Scholar]
- 7.Miyazaki M., Kim, Y. C. & Ntambi, J. M. (2001) J. Lipid Res. 42, 1018-1024. [PubMed] [Google Scholar]
- 8.Jones B. H., Maher, M. A., Banz, W. J., Zemel, M. B., Whelan, J., Smith, P. J. & Moustaid, N. (1996) Am. J. Physiol. 271, E44-E49. [DOI] [PubMed] [Google Scholar]
- 9.Pan D. A., Hulbert, A. J. & Storlien, L. H. (1994) J. Nutr. 124, 1555-1565. [DOI] [PubMed] [Google Scholar]
- 10.Enser M. (1975) Biochem. J. 148, 551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khoo D. E., Fermor, B., Miller, J., Wood, C. B., Apostolov, K., Barker, W., Williamson, R. C. & Habib, N. A. (1991) Br. J. Cancer 63, 97-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J., Ding, S. F., Habib, N. A., Fermor, B. F., Wood, C. B. & Gilmour, R. S. (1994) Int. J. Cancer 57, 348-352. [DOI] [PubMed] [Google Scholar]
- 13.Lee W. M., Ishikawa, M. & Ahlquist, P. (2001) J. Virol. 75, 2097-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ntambi J. M., Buhrow, S. A., Kaestner, K. H., Christy, R. J., Sibley, E., Kelly, T. J., Jr. & Lane, M. D. (1988) J. Biol. Chem. 263, 17291-17300. [PubMed] [Google Scholar]
- 15.Kaestner K. H., Ntambi, J. M., Kelly, T. J., Jr. & Lane, M. D. (1989) J. Biol. Chem. 264, 14755-14761. [PubMed] [Google Scholar]
- 16.Mihara K. (1990) J. Biochem. (Tokyo) 108, 1022-1029. [DOI] [PubMed] [Google Scholar]
- 17.Zheng Y., Eilertsen, K. J., Ge, L., Zhang, L., Sundberg, J. P., Prouty, S. M., Stenn, K. S. & Parimoo, S. (1999) Nat. Genet. 23, 268-270. [DOI] [PubMed] [Google Scholar]
- 18.Zheng Y., Prouty, S. M., Harmon, A., Sundberg, J. P., Stenn, K. S. & Parimoo, S. (2001) Genomics 71, 182-191. [DOI] [PubMed] [Google Scholar]
- 19.Zhang L., Ge, L., Parimoo, S., Stenn, K. & Prouty, S. M. (1999) Biochem. J. 340, 255-264. [PMC free article] [PubMed] [Google Scholar]
- 20.Miyazaki M., Kim, Y. C., Gray-Keller, M. P., Attie, A. D. & Ntambi, J. M. (2000) J. Biol. Chem. 275, 30132-30138. [DOI] [PubMed] [Google Scholar]
- 21.Stoehr J. P., Nadler, S. T., Schueler, K. L., Rabaglia, M. E., Yandell, B. S., Metz, S. A. & Attie, A. D. (2000) Diabetes 49, 1946-1954. [DOI] [PubMed] [Google Scholar]
- 22.Lo H. C., Hirvonen, M. D., Kritsch, K. R., Keesey, R. E. & Ney, D. M. (1997) Am. J. Clin. Nutr. 65, 1384-1390. [DOI] [PubMed] [Google Scholar]
- 23.Bernlohr D. A., Bolanowski, M. A., Kelly, T. J., Jr. & Lane, M. D. (1985) J. Biol. Chem. 260, 5563-5567. [PubMed] [Google Scholar]
- 24.Newton M. A., Kendziorski, C. M., Richmond, C. S., Blattner, F. R. & Tsui, K. W. (2001) J. Comput. Biol. 37, 37-52. [DOI] [PubMed] [Google Scholar]
- 25.Li C. & Wong, W. H. (2001) Proc. Natl. Acad. Sci. USA 98, 31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imanaka T., Aihara, K., Suzuki, Y., Yokota, S. & Osumi, T. (2000) Cell. Biochem. Biophys. 32, 131-138. [DOI] [PubMed] [Google Scholar]
- 27.Kersten S., Seydoux, J., Peters, J. M., Gonzalez, F. J., Desvergne, B. & Wahli, W. (1999) J. Clin. Invest. 103, 1489-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kersten S., Mandard, S., Tan, N. S., Escher, P., Metzger, D., Chambon, P., Gonzalez, F. J., Desvergne, B. & Wahli, W. (2000) J. Biol. Chem. 275, 28488-28493. [DOI] [PubMed] [Google Scholar]
- 29.Brown M. S. & Goldstein, J. L. (1997) Cell 89, 331-340. [DOI] [PubMed] [Google Scholar]
- 30.Shimomura I., Bashmakov, Y., Ikemoto, S., Horton, J. D., Brown, M. S. & Goldstein, J. L. (1999) Proc. Natl. Acad. Sci. USA 96, 13656-13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang S. L., Du, E. Z., Martin, T. D. & Davis, R. A. (1997) J. Biol. Chem. 272, 19351-19358. [DOI] [PubMed] [Google Scholar]
- 32.Cohen P., Miyazaki, M., Socci, N. D., Hagge-Greenberg, A., Liedtke, W., Soukas, A. A., Sharma, R., Hudgins, L. C., Ntambi, J. M. & Friedman, J. M. (2002) Science 297, 240-243. [DOI] [PubMed] [Google Scholar]
- 33.Peterfy M., Phan, J., Xu, P. & Reue, K. (2001) Nat Genet 27, 121-124. [DOI] [PubMed] [Google Scholar]
- 34.Moitra J., Mason, M. M., Olive, M., Krylov, D., Gavrilova, O., Marcus-Samuels, B., Feigenbaum, L., Lee, E., Aoyama, T., Eckhaus, M., et al. (1998) Genes Dev. 12, 3168-3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimomura I., Hammer, R. E., Richardson, J. A., Ikemoto, S., Bashmakov, Y., Goldstein, J. L. & Brown, M. S. (1998) Genes Dev. 12, 3182-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burant C. F., Sreenan, S., Hirano, K., Tai, T. A., Lohmiller, J., Lukens, J., Davidson, N. O., Ross, S. & Graves, R. A. (1997) J. Clin. Invest. 100, 2900-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamauchi T., Oike, Y., Kamon, J., Waki, H., Komeda, K., Tsuchida, A., Date, Y., Li, M. X., Miki, H., Akanuma, Y., et al. (2002) Nat. Genet. 30, 221-226. [DOI] [PubMed] [Google Scholar]
- 38.Soukas A., Cohen, P., Socci, N. D. & Friedman, J. M. (2000) Genes Dev. 14, 963-980. [PMC free article] [PubMed] [Google Scholar]