Abstract
Numerous bacterial pathogens use type IV secretion systems (T4SS) to deliver virulence factors directly to the cytoplasm of plant, animal, and human host cells. Here, evidence for interactions among components of the Agrobacterium tumefaciens vir-encoded T4SS is presented. The results derive from a high-resolution yeast two-hybrid assay, in which a library of small peptide domains of T4SS components was screened for interactions. The use of small peptides overcomes problems associated with assaying for interactions involving membrane-associated proteins. We established interactions between VirB11 (an inner membrane pore-forming protein), VirB9 (a periplasmic protein), and VirB7 (an outer membrane-associated lipoprotein and putative pilus component). We provide evidence for an interaction pathway, among conserved members of a T4SS, spanning the A. tumefaciens envelope and including a potential pore protein. In addition, we have determined interactions between VirB1 (a lytic transglycosylase likely involved in the local remodeling of the peptidoglycan) and primarily VirB8, but also VirB4, VirB10, and VirB11 (proteins likely to assemble the core structure of the T4SS). VirB4 interacts with VirB8, VirB10, and VirB11, also establishing a connection to the core components. The identification of these interactions suggests a model for assembly of the T4SS.
Pathogenic bacteria have evolved several strategies to subvert host cell defenses; among them is secretion of virulence factors into the host cell environment or, more craftily, directly to the interior of the host cell. This task requires the transport of macromolecules across both the bacterial envelope and the plasma membrane of the host cell. Bacterial secretion systems are classified into several subgroups, types I–V (1–5), and share evolutionarily related components. Type III and type IV secretion systems allow transport to the host cytoplasm and are related to flagellar core components (type III) and conjugation machines (type IV) (4). Type IV secretion systems (T4SS) comprise a class of transporters remarkably diverse in both the variety of substrates transferred and their promiscuity with regard to host cell types.
T4SS deliver virulence factors from Gram-negative bacterial pathogens to the cytoplasm of plant, animal, and human host cells (4, 6–8), where they enable the bacterium to modify and/or evade host cell defenses. Several T4SS are uniquely able to transfer DNA as well as proteins. Bacterial conjugation is a T4SS-mediated process responsible for genetic exchange between bacteria and contributes to the dissemination of antibiotic resistance in bacterial populations (9). Genomic sequencing efforts continue to expand the list of bacterial pathogens harboring T4SS, which now includes Agrobacterium tumefaciens, Bartonella henselae, Bordetella pertussis, several Brucella species (suis, abortus, melitensis), Campylobacter jejuni, Coxiella burnetii, Helicobacter pylori, Legionella pneumophila, Rickettsia prowazekii, Wolbachia spp, and several others (4, 6, 7, 10). T4SSs are specifically required for virulence in many of these pathogens.
Current understanding of the structure and function of T4SS derives largely from A. tumefaciens, a soil-borne pathogen that genetically transforms plants and manifests disease in nature as crown gall tumors (11). Virulence functions, including the T4SS, are encoded by the vir genes of the tumor-inducing (Ti) plasmid. A single-stranded segment of DNA (T strand) is transferred from the Ti plasmid to the plant cell, where it integrates into the host genome and directs expression of genes that result in tumor formation. The process is similar to conjugal DNA transfer; the endonuclease, VirD2, nicks and remains covalently coupled to the 5′ end of the T strand and is likely the substrate recognized by the T4SS. The similarity to plasmid conjugation is supported by the ability of the T4SS to transfer exogenous mobilizable plasmids to recipient bacteria and plants (12, 13). Proteinaceous virulence factors, VirE2 and VirF, are also secreted to the plant cell cytoplasm (14). Researchers capitalize upon A. tumefaciens' natural capacity for genetic engineering by replacing T-DNA sequences with exogenous DNA of interest.
The 11 genes of the A. tumefaciens virB operon (virB1–virB11) and the virD4 gene encode the necessary components of the T4SS. Several lines of evidence suggest a subset of the encoded products, VirB7–VirB10, assemble the structural core of the transporter. Notably, the stability of many of the VirB proteins depends on synthesis of the core components VirB7 and VirB9 (15, 16). Also, detergent extraction from membrane preparations evidences a high molecular weight complex containing VirB8, VirB9, and VirB10 (17). VirB8, VirB9, and VirB10 physically interact (18), and VirB8 is critical for recruitment of VirB9 and VirB10 to presumed T4SS assembly sites (19).
Only subsets of T4SS homologs are conserved in some bacterial species (4, 6). H. pylori, for example encodes two distinct T4SS systems; cag gene products are required for translocation of the virulence factor CagA, and com gene products promote DNA uptake and natural competence (20). com genes include homologs of virB4 and virB7–10, while cag genes include virB4, virB7–11, and virD4. In the C. jejuni pVir plasmid, an operon contributing to DNA uptake shares homology to virB8–11 (21). Finally, conjugation experiments in A. tumefaciens suggest that expression of virB7–10 in recipient A. tumefaciens strains is sufficient to stimulate recipient activity in conjugal transfer between A. tumefaciens strains (ref. 22 and A. Binns, personal communication). Thus, VirB7–VirB10 form a structural core as well as a functional core.
The most readily observed component of the A. tumefaciens T4SS is the external pilus composed primarily of VirB2 pilin subunits (23). All 11 virB genes are required for elaboration of the pilus, which presumably facilitates attachment to the host cell. The role, if any, of the pilus in mediating substrate delivery is unknown. VirB5 and homodimers of the lipoprotein, VirB7, cofractionate in pilus preparations and contribute to pilus assembly (24, 25). The F-plasmid conjugation system also elaborates a pilus. TraV is required for F-pilus biogenesis and may be functionally analogous to VirB7 (26). TraV anchors a membrane-spanning complex that includes TraV, TraK, and TraB, which are outer membrane, periplasmic, and inner membrane proteins, respectively, to the outer membrane (26). Similarly, VirB7 may anchor components required for T-pilus assembly to the outer membrane.
The peptidoglycan layer separates the inner and outer membranes and provides integrity to the cell envelope. This layer is not porous enough to allow penetration by large protein complexes and poses a significant obstacle to assembly of macromolecular complexes (27, 28). VirB1 has sequence similarity to lytic transglycosylases and likely contributes to virulence by locally disrupting the peptidoglycan, facilitating T4SS assembly (29).
VirB4, VirB6, VirB11, and VirD4 all are proposed to form pore structures in the bacterial envelope (6, 8, 11, 30). Crystal structures for VirB11 and VirD4 homologs, HP0525 and TrwB, respectively, have been solved (31, 32). Both are predicted to be hexameric ring structures embedded in the inner membrane. VirB11 and its homologs may function to traffic unfolded substrates across the cytoplasmic membrane, contributing either to secretion or T4SS assembly (32–36). ATP binding may regulate membrane affinity (37), coordinate assembly of the hexamer, or mediate interactions between the N- and C-terminal domains (32, 37). The crystal structure of H. pylori HP0525 reveals a 10-nm pore, which could facilitate transport of large molecules (32). In support, VirB11 ATPase activity is correlated with substrate transfer (36, 38), and specific VirB11 mutants are defective in elaboration of the T-pilus (36). TrwB resembles ring helicases, indicating a role in DNA transfer (31). VirB4 forms dimers, and its NTP binding may supply energy for T4SS assembly or substrate transport (39). VirB4 also affects the membrane localization of VirB3 (40). VirB6 is anchored in the inner membrane and contains multiple membrane-spanning domains (30). Strains lacking VirB6 exhibit reduced accumulation of VirB3, VirB5, and VirB7 homodimers (41), and VirB6 may influence the membrane association of VirB5 and VirB7 (17), suggesting a role in pilus assembly.
As a prelude to understanding the architecture and function of T4SS, it is critical to determine the network of protein interactions contributing to its assembly. Here, we determine interactions among VirB components by using a high-resolution yeast two-hybrid prey library and further define the minimal domains required for previously known interactions. Centromere-based yeast two-hybrid vectors were developed to facilitate library-based screening of peptide interactions, simplify analysis of potential protein interactions, and reduce the effects of expression of toxic peptides (42). Our study links the cytoplasmically exposed inner membrane protein, VirB11, a potential pore protein, with other members of the proposed core complex, VirB9 and VirB7, involved in transporter and pilus assembly, thereby establishing a link between the cytoplasm and the exterior pilus. Additional data suggest that VirB1 peptidoglycanase activity is directed to sites of T4SS assembly via interaction with several core components, primarily VirB8, but also VirB4, VirB9, VirB10, and VirB11. VirB4 interactions with VirB8, VirB10, and VirB11 establish its association with core components. These results, coupled with previous findings, suggest a model for assembly of T4SS.
Materials and Methods
Strains and Culture Conditions.
Standard molecular protocols were followed with little modification. Escherichia coli strains DH5α and JA226, as well as yeast strain YD116 (42), were grown in Luria broth, M9 minimal media lacking leucine, and yeast synthetic minimal media lacking leucine, tryptophan, and/or uracil as appropriate. Yeast two-hybrid vectors were derived from pCD.1, pC-ACT.1, pCD.2, and pC-ACT.2 (42). DNA was introduced into E. coli either by the method of Inoue et al. (43) or electroporation and introduced into yeast by the lithium acetate method (44). DNA isolated from E. coli strain JA226 was phenol-extracted to eliminate nuclease activity. DNA for sequencing was prepared by using Qiagen spin columns. Sequencing was performed on an ABI377 with a BigDye terminator cycle sequencing kit (Applied Biosystems). Source DNA for plasmid and library construction included A. tumefaciens pTiC58; pGK217, a pUC119 derivative containing the virB operon (45); and pMTX120, a pACYC derivative with a 2.7-kb EcoRI fragment that expresses VirD4 from a plac promoter (M. Llosa, personal communication). PCR amplification was used to provide appropriate fragments for cloning.
Construction of High-Resolution Prey Library.
pC-ACT.1 (42) was modified to create pC-ACT.3. The sequence, inserted between BamHI and PstI sites (in parentheses), (GGATCC)GTGCACCTGCAGGACTAG(TTGCAG), creates an SpeI site (underlined) and eliminates the PstI site (in bold). Equal molar amounts of pGK217 and pMTX120 were mechanically sheared and size-selected to a mean fragment size of 500–700 bp. DNA linkers, 5′-(CTAG)ACCTAGGTACC[-/T/TC], were ligated to the sheared DNA. The linker contains an SpeI compatible overhang (in parentheses). The 3′ blunt end was constructed in three frames (in brackets) (i.e., three equal molar linker populations each differing in length by 1 bp) to facilitate creation of in-frame fusion junctions. The sheared DNA+linker was ligated to pC-ACT.3 in the presence of SpeI and transformed into E. coli strain DH5α. Upon ligation, the SpeI site (in bold) is destroyed.
The prey library consists of ≈100,000 independent transformants and contains fusions at every nucleotide position in the vir-coding sequences with >90% certainty as calculated by using the equation n = ln(1 − P)/ln(1 − I/S), where P = probability of representation, n = 100,000 transformants, I = 1 bp (size of target fusion sequence is 1 bp), and S = 34,000 bp {size of potential target population is [12,700 bp (pGK217) + 4,300 bp (pMTX120)] × 2 orientations = 34,000 bp}.
Construction of Bait and Prey Vectors.
Identical peptides were cloned into the polylinker region of pCD.1 or pCD.2 (bait) and pC-ACT.1 or pC-ACT.2 (prey). The peptides used for assaying potential pairwise interactions among VirB proteins are indicated as follows, with the numbers in parentheses indicating the amino acid residues included in the two-hybrid constructs: VirB1 (1–252), VirB2 (1–121), VirB2 (47–121), VirB3 (3–108), VirB5 (2–95), VirB6 (3–295), VirB7 (3–55), VirB8 (2–237), VirB9 (3–292), VirB10 (3–337), and VirB11 (1–344). With the exception of VirB2 and VirB5, all are essentially full-length peptides. All baits except VirB2 and VirB10 were used to screen the high-resolution library. Several baits did not retrieve prey from the library.
Library Screening.
Yeast containing the bait of interest were transformed with the prey library. A typical transformation represented 2 × 106 transformation events, ensuring 20-fold representation of the library. Potential interaction clones were selected on appropriate minimal media (42). For secondary confirmation, clones that grew on selective media were restreaked to selective media and visually screened for β-galactosidase activity as described (42). To reconfirm interactions, prey plasmid DNA was extracted from isolates exhibiting strong β-galactosidase activity and cotransformed with the bait plasmid into YD116 and tested for growth on selective media. Candidate prey plasmids were sequenced. Some isolates, categorized as false positives, did not contain in-frame fusions to vir coding sequences. The most prevalent class (≈50%) contained partial sequence of the chloramphenicol acetyltransferase gene from pMTX120. The remainder of false positives included inverted, out-of-frame fusions or vir coding sequences with rearrangements or insertions and are not presented in the results.
Results
Interactions between most of the nearly full-length VirB proteins were first examined. Of pairwise combinations tested, positive interactions were observed only between VirB7 and VirB9 and, curiously, only with VirB7 bait and VirB9 prey. Lack of reciprocal interactions has been previously observed with more traditional two-hybrid vectors based on the 2-μ origin of replication (46). The paucity of interactions was surprising given existing evidence for T4SS protein–protein interactions. Sometimes, expected peptide interactions are not observed by the two-hybrid assay because of improper folding or decreased protein stability of particular fusion proteins (47).
The components of the A. tumefaciens T4SS are largely membrane associated. Potentially, the nearly full-length hydrophobic proteins included in our constructs are unable to fold properly in the hydrophilic yeast cytoplasm, or perhaps only a limited number of peptide regions are suitable for interaction. Thus, we reasoned that small peptide domains may have a greater probability of adopting a native conformation in the yeast cytoplasm, permitting detection of predicted two-hybrid interactions. Because of the size of the virB coding sequence (≈9.5 kb), a prey library of sufficient resolution could be feasibly constructed to represent all possible insert fusion junctions and encode a correspondingly complex set of potential small peptide domains. To this end, we constructed a high-resolution prey library containing the entire virB operon and virD4.
Existing bait constructs (see Materials and Methods) were used to screen the high-resolution prey library. Several baits, which did not exhibit interactions in pairwise combinations with full-length prey, were able to retrieve interacting peptide prey from the library. Importantly, peptides originally isolated as prey were observed to be more effective baits as compared with their full-length protein counterparts.
High-Resolution Two-Hybrid Library Approach Details Extent of Peptide Interaction Domains.
Nearly full-length VirB7, VirB7(3–55), retrieved prey efficiently from the library, whereas VirB9(3–293) did not retrieve any prey from the library. This result is similar to the directionality of the full-length, pairwise interactions we observed. DNA from 62 prey isolated with VirB7(3–55) bait was sequenced and found to contain in-frame fusions to VirB9 (Fig. 1A). Fusions occurred at 33 unique VirB9 amino acid residues, demonstrating the resolution of the library.
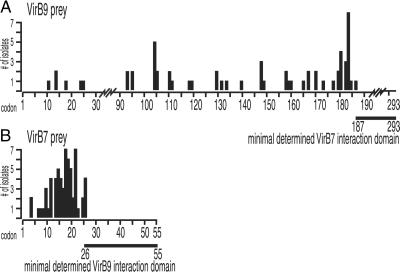
Fig 1.
Minimal interacting domains of VirB7 and VirB9 proteins. The N-terminal residues of VirB9 and VirB7 peptide prey isolated from the high-resolution library were determined. The position of the peptide termini are plotted versus the codon position of the full-length prey protein (x axis) and the number of isolates identified (y axis). The lines below each plot represent the minimal prey domain determined to interact with the bait used. (A) VirB9 prey isolated with full-length VirB7 bait. (B) Total VirB7 prey isolated with two VirB9 baits, VirB9(14–293) and VirB9(183–293).
A plot of fusion junctions versus VirB9 amino acid sequence reveals several interesting features (Fig. 1A). Because only sequences C-terminal to the VirB9 fusion junction are translated in the fusion product, the most C-terminal fusion junction isolated delimits the maximal N-terminal truncation that allows interaction. Therefore, the smallest VirB9 peptide that interacts with the VirB7 bait is 107 aa in length, VirB9(186–293) (Fig. 1A). This result is in close agreement with previous studies showing VirB9 residues 173–275 interact with VirB7 (48). Therefore, the minimal domain of VirB9 able to interact with VirB7 is likely in the 90-residue region between amino acids 186 and 275.
The majority of fusion junctions occurred between VirB9 residues 94 and 186. No fusion junctions were identified between residues 25 and 94. This finding may suggest peptide fusion junctions that are permissive (94–186) or nonpermissive (25–94) for two-hybrid interaction. Nonpermissive fusion junctions may represent peptides that genuinely do not interact with the bait or peptides that fold improperly, are unstable, or are underrepresented in the library, perhaps because of toxicity.
Peptide Domains Exhibit Interactions Not Observed with Full-Length Baits.
The above results demonstrate that a high-resolution library can identify interacting peptides and delimit the minimal C-terminal regions required for interaction. Because full-length VirB9 was not effective as a bait, we tested VirB9 peptides, isolated as prey, as baits. We chose two VirB9 prey peptides, one nearly full length, residues 14–293, and a small peptide domain, residues 183–293.
In contrast to full-length VirB9, the smaller peptides were successful baits. We sequenced 75 prey isolates retrieved by both VirB9 peptide baits. Of these, 70 prey were in-frame fusions to VirB7 (Fig. 1B), of which four included VirB6 coding sequence translated out-of-frame with respect to VirB6 but in-frame for VirB7. Additionally, VirB9(14–293) retrieved three prey identified as VirB1 fusions (Fig. 2B) and two prey expressing identical in-frame fusions beginning 5′ of the virB11 translation initiation codon (data not shown).
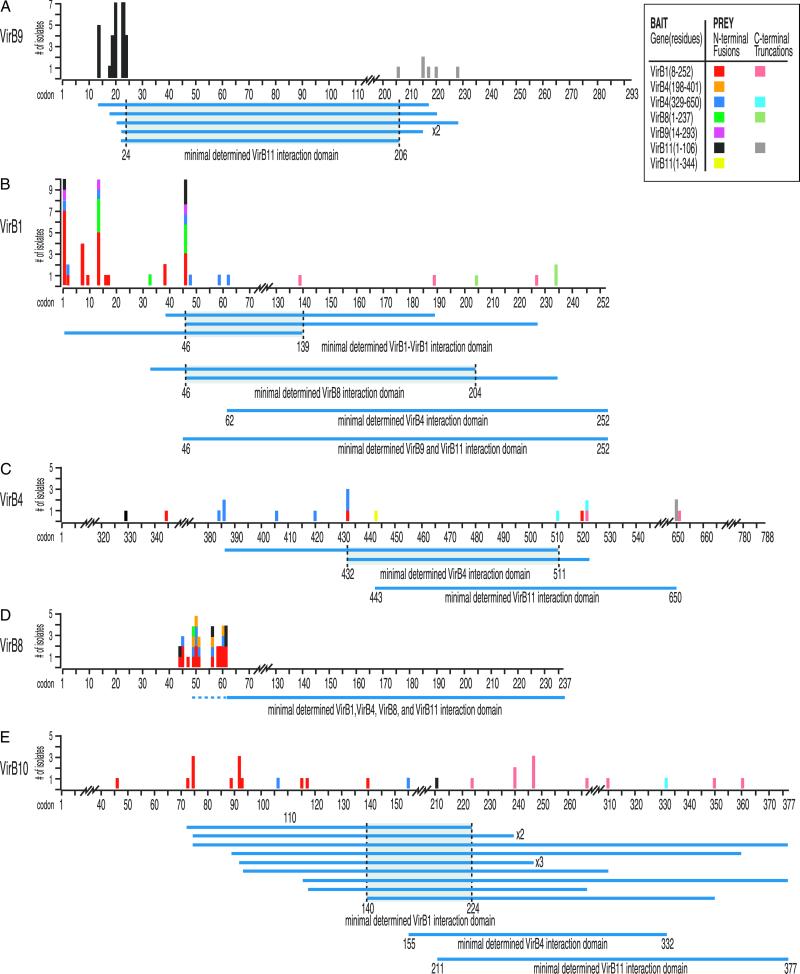
Fig 2.
Minimal interacting domains of VirB proteins. The N-terminal and C-terminal residues of prey isolated from the high-resolution library are plotted versus codon position (x axis) and the number of isolates identified (y axis). Data are color-coded for each bait used (see Inset). Prey that have determined N and C termini and that were isolated by a unique bait are aligned to define regions in common, indicating minimal interacting domains (horizontal blue lines below the amino acid numbers). The prey represented are as follows VirB9 (A), VirB1 (B), VirB4 (C), VirB8 (D), and VirB10 (E).
Thus, residues 26–55 of VirB7 constitute the minimal determined domain sufficient to interact with VirB9, and VirB9 residues 187–293 are sufficient to interact with VirB7 (Figs. 1 and 3). This reciprocity argues that the observed interactions reflect in vivo interactions. As the peptide VirB9(14–293) contains an N-terminal VirB9 sequence not included in the VirB9(183–293) peptide, these residues (14–182) may contain VirB1 and VirB11 interaction domains. Indeed, this prediction is supported by the results below (Fig. 3).
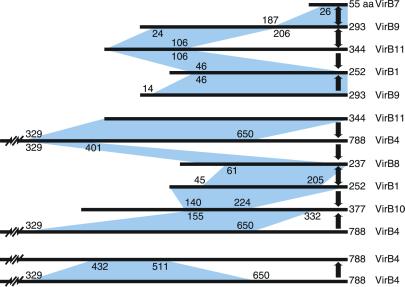
Fig 3.
VirB protein interactions. The pairwise interactions identified among the VirB proteins are summarized. The horizontal lines represent the peptide sequence of each VirB protein with the length in amino acids indicated to the right. Blue shaded regions indicate the minimal domains that contribute to each pairwise interaction. The numbers indicate the specific residue in the coding sequence that delimits each domain. The heavy arrows indicate the direction in which the interaction was observed, from bait to prey. The data most easily sort into two main complexes; the first consists of VirB1, VirB7, VirB9, and VirB11, and the second consists of VirB1, VirB4, VirB8, VirB10, and VirB11. VirB4–VirB4 interactions are presented as a third group. VirB1–VirB1, VirB1–VirB4, VirB8–VirB8, VirB8–VirB10, and VirB10–VirB11 interactions are not represented but are addressed in the text.
The contrast between the success of VirB9 peptide fragments in retrieving prey from the library and the lack of prey retrieved by full-length VirB9 suggests that identifying appropriate baits is a significant limitation when screening two-hybrid libraries. Although VirB9 peptides obtained as prey were effective as baits, this was not true of all prey peptides tested. Fig. 2 presents interaction data obtained with effective baits. The observed interactions include bait and prey peptides of VirB1, VirB4, VirB8, VirB9, VirB10, and VirB11. These interactions are presented below, organized by potential functional protein groups. Fig. 2 presents the data sorted by the prey identified. Most prey were identified by more than one bait (e.g., VirB1 prey were retrieved by VirB1, VirB4, VirB8, VirB9, and VirB11 baits). Data from the baits that retrieved each prey are color-coded and plotted with respect to each prey protein (Fig. 2).
VirB11, an Inner Membrane, Potential Pore-Forming Protein Interacts with Periplasmic VirB9.
The crystal structure of the VirB11 homolog, HP0525 of H. pylori, has two major domains: an N-terminal membrane-associated domain, predicted to extend into the periplasmic space, and a C-terminal cytoplasmic domain (32). VirB11 may facilitate substrate transport across the inner membrane, and VirB11 interactions are likely to reflect basal components of T4SS architecture. We constructed bait plasmids expressing the N-terminal membrane-associated residues VirB11(1–106) and the C-terminal cytoplasmic residues VirB11(107–344).
The N-terminal domain retrieved many prey from the library, whereas the C-terminal domain did not retrieve any. Thirty-seven prey retrieved identified VirB1(4), VirB4(1), VirB8(4), VirB9(29), and VirB10(1) (Fig. 2 B, C, D, A, and E, respectively). Additional sequencing revealed six of the 29 VirB9 prey were also C-terminally truncated, further refining a minimal interacting domain within VirB9 (Figs. 2A and 3). Full-length VirB11(1–344) retrieved two identical VirB4 prey peptides spanning residues 329–650 (Figs. 2C and 3). Interestingly, the VirB11 N-terminal domain retrieved prey that are also periplasmic or extend into the periplasm.
VirB9 Has at Least Two Discrete Interaction Domains and Interacts with VirB1, VirB7, and VirB11.
With the exception of two VirB11 prey isolated with bait peptide VirB9(14–293), the observed VirB9–VirB11 two-hybrid interactions are largely unidirectional (i.e., VirB11 bait identifies numerous VirB9 prey). When the N- and C-terminal sequences of VirB9 prey are examined, the smallest VirB9 domain required for interaction with VirB11 lies between VirB9 residues 24 and 206 (Fig. 2A). Thus, VirB9 seems to have two interaction domains, an N-terminal domain that interacts with the N-terminal domain of VirB11, and a C-terminal domain that interacts with VirB7 (Fig. 3).
Additional observed interactions of VirB9 and VirB11 baits with VirB1 prey are also unidirectional (Fig. 3 and below) and did not significantly define a minimal interaction domain within the VirB1 peptide (Figs. 2B and 3). Vectors expressing the C-terminal portion of VirB1 are toxic in yeast, even in the low copy number CEN-based vectors (C. Hackworth and P.C.Z., unpublished results). This finding may explain our inability to narrowly define an interaction domain within the VirB1 protein. However, the C terminus of VirB1, VirB1*, associates with VirB9–VirB7 heterodimers by chemical cross-linking and coimmunoprecipitation (49). Our results are consistent with this interaction. Minimal domains of VirB4 (Fig. 2C), VirB8 (Fig. 2D), and VirB10 (Fig. 2E) prey that interact with VirB11 were also determined. Consistently, the domains of all prey identified are predicted to be periplasmically exposed (30, 50).
VirB4 Peptides Interact with VirB1, VirB4, VirB8, and VirB10.
Full-length VirB11 retrieved a VirB4 peptide spanning residues 329–650. This peptide was cloned and used as a bait to screen the library. In addition, we constructed a VirB4 bait containing residues 198–401. Both VirB4 peptides retrieved a total of 12 (5 + 7) VirB8 prey (Fig. 2D). Additionally, VirB4(329–650) retrieved VirB1(7), VirB4(7), and VirB10(2) prey (Fig. 2 B, C, and E, respectively). The peptide VirB4(329–650) is predicted to span a periplasmic domain, consistent with interactions including prey domains that are predicted to be periplasmic (50) (see Discussion). Whereas secondary structure analysis predicts VirB4 residues 198–401 are cytoplasmically exposed (50), our findings of VirB4(198–401) interaction with a primarily periplasmic domain of VirB8 suggest this VirB4 domain may adopt a membrane or periplasmic conformation when incorporated into a complex with other T4SS components.
VirB1 Exhibits Multiple Interactions with Peptides of VirB1, VirB4, VirB8, and VirB10.
VirB1 has sequence similarity to lytic transglycosylases and may locally remodel the peptidoglycan to provide a channel for transporter assembly (29). Therefore, it is reasonable to expect VirB1 to interact with core components of the transporter to locally direct VirB1 activity. A full-length VirB1 bait, VirB1(1–252), did not interact with other full-length constructs in our pairwise screen. VirB1, however, successfully retrieved prey from the high-resolution library. Of 56 isolates sequenced, in-frame fusions to VirB1(23), VirB4(3), VirB8(17), and VirB10(13) were identified (Fig. 2). As noted above, VirB9 and VirB11 prey were not identified in the isolates sequenced.
As with the VirB7–VirB9, VirB1–VirB4, and VirB9–VirB11 two-hybrid interactions (above), reciprocal interactions argue that the interactions revealed reflect in vivo interactions. Nearly full-length VirB8(2–237) was a less effective bait; of eight prey sequenced, seven were in-frame fusions to VirB1 and one was VirB8 (Fig. 2).
C-Terminal Analysis of Prey Peptides Further Narrows Interaction Domains.
The library, by design, is biased toward generation of N-terminally truncated prey. However, isolated prey may express peptides that are truncated at the C terminus if the inserted fragment is not sufficiently large to contain the entire C-terminal coding sequence. Restriction analysis indicated several prey with possible C-terminal truncations that were confirmed by sequencing (Fig. 2). Complete C-terminal coding sequences are not indicated. Prey with determined N and C termini were aligned to define regions common to prey isolated with a unique bait (horizontal blue lines below the amino acid numbers, Fig. 2) to determine minimal interacting domains. VirB10 residues 140–224 delimit the VirB1 interaction domain, and VirB10 residues 155–332 delimit the VirB4 interaction domain (Fig. 2E). Interaction domains within VirB1 were also delimited to residues 46–139 for VirB1–VirB1 interactions, and residues 46–204 for interaction with VirB8 (Fig. 2B). VirB4–VirB4 interactions were delimited to residues 432–511 (Fig. 2C).
In summary, we have observed reciprocal interactions with VirB7 and VirB9, VirB9 and VirB11, VirB1 and VirB8, VirB1 and VirB4, and among VirB4 fragments (Fig. 3). Additionally, we have evidence for VirB1–VirB1 and VirB8–VirB8 interactions. Finally, VirB8 was isolated with two distinct but overlapping VirB4 peptide baits, VirB4(329–650) and VirB4(198–401). From this, we infer that VirB4 residues 329–401 delimit a VirB8 interaction domain (Fig. 3). VirB11(1–344) and VirB11(1–106) also retrieved VirB4 prey and delimit VirB4 residues 443–650 as a VirB11 interaction domain.
Discussion
Construction of a high-resolution two-hybrid library expressing small peptide domains of the 11 VirB proteins and VirD4 enabled identification of several VirB protein interactions. Significantly, interactions between VirB11 and VirB8, VirB9, and VirB10 and between VirB4 and VirB8 and VirB10 provides evidence for complete T4SS interaction pathways linking the cytoplasmic components to the external pilus (Fig. 4). The putative lytic transglycosylase, VirB1, interacts with potential core proteins VirB4, VirB8, VirB9, VirB10, and VirB11. The VirB1–VirB8 interaction may further implicate VirB8 in the initial establishment of a T4SS assembly site, locally directing VirB1 activity and facilitating the recruitment of other Vir proteins into a stable complex (Fig. 4). Besides identifying putative protein–protein interactions, we significantly defined the domains involved, demonstrating the utility of a high-resolution library for narrowly determining interaction domains (Fig. 3), particularly among members of membrane-associated complexes. In several instances, peptides isolated as prey were more successful baits than their nearly full-length counterparts. Interestingly, all of the interactions described involve peptide domains predicted to be exposed to the periplasm. The described interactions may reflect transient interactions in an assembly pathway or stable interactions in a completed structure. Confirmation of interactions detected by our high-resolution two-hybrid approach awaits further biochemical studies. However, the revealed interactions can focus the direction of future investigation and stimulate the generation of T4SS assembly or structure hypotheses.
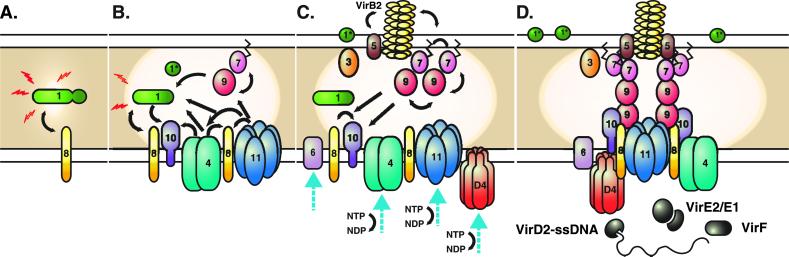
Fig 4.
Model for assembly of the Agrobacterium T4SS. Protein interactions determined in this work are depicted in A and B. Previously determined interactions are shown in C. The double lines at the top and bottom of each panel represent the inner and outer membranes, and the shaded region represents the periplasmic space and peptidoglycan. (A) VirB8 functions as a “founding member” of the T4SS and recruits VirB1 to the site of assembly. VirB1 locally remodels the peptidoglycan (represented by decreased shading of the periplasm). (B) VirB1 activity allows recruitment of other T4SS components such as VirB4, VirB7, VirB9, VirB10, and VirB11 by clearing the peptidoglycan or by recruiting components via direct interactions. VirB7–VirB9 heterodimers are critical for stability of T4SS proteins. (C) As the assembly matures, the remaining components are recruited, including VirB3, the pilus (VirB2, VirB5), and inner membrane components (VirB6, VirD4). VirB1* may function extracellularly, and its loose association with the surface is indicated. VirB7 and VirB5 contribute to pilus assembly. VirB4, VirB6, VirB11, and VirD4 all have been postulated to create channels for substrate secretion (blue arrows). (D) A model for assembled T4SS based on interactions described in A–C. VirB7–11 and VirB4 form a functional core of the T4SS that spans both bacterial membranes. Known substrates for export include VirE2 and, perhaps, its specific chaperone VirE1, VirF, and the T-complex (VirD2–ssDNA). VirD4 is likely the recognition protein coupling VirD2 export to the T4SS and may function as a DNA-helicase to liberate the T-strand from the Ti plasmid. VirB11 forms a pore and may facilitate substrate export.
VirB11 and VirB9 Interactions.
The data presented provide evidence completing an interaction pathway between VirB11 and pilus subunit, VirB2, via VirB9 (Figs. 3 and 4). VirB11 previously has not been shown to physically interact with other T4SS components. We also define two discrete interaction domains within VirB9: an N-terminal domain, VirB9(24–206), which interacts with VirB11, and a C-terminal domain, VirB9(187–293), which interacts with VirB7. Recently, VirB7 has been shown to copurify with exocellular pilus components, VirB2 and VirB5 (24). Thus, the interaction pathway is VirB11-VirB9-VirB7-VirB2/VirB5 (Fig. 4). That specific mutations in VirB11 impair pilus biogenesis (36) provides genetic evidence that VirB11 is properly placed in this physical interaction pathway.
Previously, VirB9 was shown to interact with VirB7 (46, 48, 51, 52), as well as itself, VirB8, and VirB10 (18). The VirB9 peptide, VirB9(17–122), was shown to interact with VirB8, VirB9, and VirB10 and the peptide VirB9(173–295) with VirB7 (18, 48). When evaluated with our results, the VirB9 peptide domain supporting VirB7 interaction can be further limited to residues 187–295. The VirB9 domain that supports VirB11 interaction spans residues 24–206 and includes most of the region supporting interaction with VirB8, VirB9, and VirB10. Similarly, the peptide VirB11(1–106) also interacts with VirB8, VirB9, and VirB10; it is not known whether VirB11 subdomains exist that specifically interact with VirB8, VirB9, or VirB10.
The formation of VirB7–VirB9 heterodimers stabilizes VirB11, and VirB11 stability is reduced in the absence of either VirB7 or VirB9 (16). Our data suggest that VirB11 stability is promoted by a VirB9–VirB11 interaction. Stabilization, however, may result from indirect interactions. For example, VirB10 abundance is also reduced in the absence of VirB7 and VirB9 (52). Thus, VirB7–VirB9 heterodimers may influence VirB11 stability indirectly via interactions with VirB10. Overexpression of either VirB9 or VirB10 results in increased abundance of VirB11 (53). Thus, although we did not detect a VirB10–VirB11 interaction, both VirB9 and VirB10 may interact with VirB11 to confer stability.
VirB4 Interactions.
VirB4 interacts with itself, VirB8, VirB10, and VirB11 in our assay. As with VirB11, VirB4 stability depends on VirB7–VirB9 heterodimers that may promote VirB4 stability directly or indirectly via stabilization of VirB10 (52). The VirB4–VirB10 interaction suggests direct causality. VirB6 also influences VirB4 stability (39), yet we did not detect VirB6 interactions. The significantly hydrophobic nature of the VirB6 protein may preclude the representation of suitable domains for interaction in our library. The VirB4–VirB8 interaction observed is intriguing because it suggests that VirB4 ATPase activity may contribute to early T4SS assembly steps (Fig. 4). However, VirB4–VirB10 and VirB4–VirB11 interactions could indicate a more structural involvement in transporter activity.
Both VirB4 and VirB11 are cytoplasmically exposed (32, 37, 50) and have ATPase activity, which may provide energy for assembly of the T4SS or for substrate translocation (4). VirB4 has four potential transmembrane regions, suggesting a topology with two periplasmic domains, spanning residues 37–108 and 464–608. The second periplasmic domain, residues 464–608, is flanked by Walker A and B motifs, which are proposed to come together, forming a periplasmic loop that may interact with other integral membrane components of the T4SS (50). Interaction of peptide VirB11(1–106) with the VirB4 domain (443–650) spanning the VirB4 periplasmic loop correlates nicely with this prediction. The periplasmic loop of VirB4 and N-terminal domain of VirB11 both interact with VirB8 and VirB10, suggesting close association of putative pore proteins in the periplasm. However, with the baits tested, we did not detect interactions between VirB4 and VirB9.
VirB1 Interactions.
The peptidoglycan is not porous enough to allow penetration by large protein complexes (27, 28). Thus, lytic transglycosylases, such as VirB1, are proposed to provide local lysis of the peptidoglycan, permitting assembly or insertion of macromolecular complexes. Our results indicate interactions between VirB1 and VirB4, VirB8, VirB9, VirB10, and VirB11. Of these interactions, the VirB1–VirB8 interaction was reciprocal. VirB8 is predicted to contain a single N-terminal membrane-spanning region resulting in the majority of the C terminus extending into the periplasm (54). VirB1 has no membrane-spanning region but has a sec-dependent signal peptide, predicting export to the periplasm where it may act to dissolve the peptidoglycan. Interaction of VirB1 with VirB8 provides a means to spatially restrict VirB1 activity to sites of T4SS assembly and agrees with VirB1 localization to the periplasmic side of the inner membrane (55).
VirB1–VirB8 interaction is supported by comparison to the ptlE gene in B. pertussis. The B. pertussis T4SS delivers pertussis toxin to host cells and is encoded by ptlA-I. The ptl operon contains homologs of VirB proteins except VirB1 and VirB5 (4). PtlE, the VirB8 homolog, was recently shown to have peptidoglycanase activity. Whereas the C terminus of PtlE and VirB8 share homology, peptidoglycanase activity maps to the nonhomologous, N-terminal, PtlE residues 1–102 (56). Evidence for spatially restricted peptidoglycanase activity is also observed for P19, a homolog of VirB1, important for efficient conjugative transfer of plasmid R1 (28, 57). P19 overexpression results in localized cell lysis, suggesting that P19 is targeted to specific regions of the cell envelope (57).
VirB8 is required for localization of VirB9 and VirB10 to discrete sites in the bacterial membrane (19). However, the localization pattern of VirB8 is independent of other VirB protein (19), suggesting that VirB8 functions to nucleate assembly of the T4SS. Paradoxically, the formation of VirB7–VirB9 heterodimers (51) and high molecular weight VirB10 complexes (15) does not require VirB8. Potentially, T4SS subassemblies can form but may not lead to the construction of a localized T4SS structure in the absence of VirB8.
The levels of VirB4 and VirB11 are diminished in a virB1 deletion strain (58), which may reflect the VirB1–VirB4 and VirB1–VirB11 interactions we observe, or result from loss of VirB1 peptidoglycanase activity. Our data suggest VirB1 directly recruits VirB4, VirB9, and VirB11, facilitating their incorporation into a stable T4SS structure. VirB1 peptidoglycanase activity, however, supports an indirect role. By opening the peptidoglycan at specific sites, VirB1 may favor assembly of T4SS components at sites of assembly, indirectly promoting additional stabilizing interactions.
VirB1 may promote VirB11 stability by creating space within the peptidoglycan allowing VirB9–VirB11 interactions; VirB1 may be more critical if the larger VirB7–VirB9 heterodimers confer VirB11 stability. Similarly, VirB4 stability may depend on the formation of VirB10 complexes, as suggested by the observed VirB4–VirB10 interaction. VirB10 complex formation depends on VirB9 (15), and preliminary results suggest that VirB1 also contributes to the formation of high molecular weight VirB10 complexes (J.R.Z. and P.C.Z., unpublished results). Therefore, VirB1 may open the peptidoglycan, and the recruitment of VirB7–VirB9 heterodimers stabilizes VirB11 and promotes VirB10 complex formation. VirB1 also may promote association of the core components VirB4 and VirB8–11 via direct interactions.
VirB1 is proteolytically processed to liberate a C-terminal peptide, VirB1*, that is found loosely associated with the cell surface (49). Both the N-terminal VirB1 peptide and VirB1* partially restore virulence to strains lacking VirB1, suggesting separable VirB1 functions (59). Opening of the peptidoglycan is likely one activity; perhaps facilitating recruitment of other T4SS components is another. Although not strictly required for T-DNA transfer (13, 58), VirB1 is required for pilus biogenesis (60). The VirB1–VirB9 interaction, when viewed in light of the intimate association of VirB9 with pilus-associated protein VirB7, suggests VirB1 may directly contribute to pilus assembly (49).
Implications for Transporter Assembly.
Many of the interactions we observed are in agreement with existing models (4, 6). We depict an outline for transporter assembly based on interactions observed in this work (Fig. 4 A and B) and previously identified physical interactions (Fig. 4C). VirB8 likely nucleates assembly of the transporter at specific sites in the membrane (19) and may direct VirB1 peptidoglycanase activity (Fig. 4A). Opening of the peptidoglycan then facilitates recruitment of the core components. As VirB1 is required for elaboration of the pilus (60), we speculate that VirB1 may greatly facilitate T4SS assembly, but also have an essential function later in pilus biogenesis.
VirB7–VirB9 heterodimers may be recruited to assembly sites, perhaps via VirB1–VirB9 or VirB8–VirB9 interactions (Fig. 4 B and C), where they stabilize other T4SS components. VirB11 directly associates with VirB9; thus, a direct interaction pathway, VirB11-VirB9-VirB7-VirB2/VirB5, links VirB11 to the pilus. VirB4 may be linked to the transporter via interactions with VirB10 and VirB11. VirB8 associates with T4SS components including itself, VirB1, VirB4, VirB8–10 (18), and VirB11, suggesting a fundamental role in establishing T4SS architecture (Fig. 4D). Current thoughts regarding subsequent T4SS assembly steps that lead to pilus biogenesis and involve VirB2, VirB5, VirB6, and VirB7 are detailed elsewhere (17).
The present study is an attempt to use a peptide library to screen for interactions in the A. tumefaciens T4SS. The success of this approach can be pursued further with peptide baits not yet tested, allowing investigators to produce an even more detailed view of the architecture of this critical transport system. Understanding T4SS architecture will allow predictions of T4SS protein function, facilitate future research, and perhaps provide targets for development of therapeutics that disrupt T4SS to combat disease or hinder the spread of antibiotic resistance in microbial populations.
Acknowledgments
We thank P. Silverman for sharing results and for discussions contributing to this work and T. Durfee for valuable input. This work was conducted with the support of a National Science Foundation Postdoctoral Research Fellowship in Microbiology (to D.V.W.) and a grant through the Plant and Microbial Biology-Torrey Mesa Research Institute (Syngenta) Research Agreement to the Department of Plant and Microbial Biology at the University of California, Berkeley.
Abbreviations
T4SS, type IV secretion system
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on May 1, 2001.
References
- 1.Linton K. J. & Higgins, C. F. (1998) Mol. Microbiol. 28, 5-13. [DOI] [PubMed] [Google Scholar]
- 2.Sandkvist M. (2001) Mol. Microbiol. 40, 271-283. [DOI] [PubMed] [Google Scholar]
- 3.Plano G. V., Day, J. B. & Ferracci, F. (2001) Mol. Microbiol. 40, 284-293. [DOI] [PubMed] [Google Scholar]
- 4.Christie P. J. & Vogel, J. P. (2000) Trends Microbiol. 8, 354-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacob-Dubuisson F., Locht, C. & Antoine, R. (2001) Mol. Microbiol. 40, 306-313. [DOI] [PubMed] [Google Scholar]
- 6.Baron C., O'Callaghan, D. & Lanka, E. (2002) Mol. Microbiol. 43, 1359-1365. [DOI] [PubMed] [Google Scholar]
- 7.Sexton J. A. & Vogel, J. P. (2002) Traffic 3, 178-185. [DOI] [PubMed] [Google Scholar]
- 8.Christie P. J. (1997) J. Bacteriol. 179, 3085-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christie P. J. (2001) Mol. Microbiol. 40, 294-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao T. B. & Saier, M. H., Jr. (2001) Microbiology 147, 3201-3214. [DOI] [PubMed] [Google Scholar]
- 11.Zupan J., Muth, T. R., Draper, O. & Zambryski, P. (2000) Plant J. 23, 11-28. [DOI] [PubMed] [Google Scholar]
- 12.Buchanan-Wollaston V., Passiatore, J. & Cannon, F. (1987) Nature (London) 328, 172-175. [Google Scholar]
- 13.Fullner K. J. (1998) J. Bacteriol. 180, 430-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vergunst A. C., Schrammeijer, B., den Dulk-Ras, A., de Vlaam, C. M., Regensburg-Tuïnk, T. J. & Hooykaas, P. J. (2000) Science 290, 979-982. [DOI] [PubMed] [Google Scholar]
- 15.Beaupré C. E., Bohne, J., Dale, E. M. & Binns, A. N. (1997) J. Bacteriol. 179, 78-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandez D., Spudich, G. M., Zhou, X. R. & Christie, P. J. (1996) J. Bacteriol. 178, 3168-3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krall L., Wiedemann, U., Unsin, G., Weiss, S., Domke, N. & Baron, C. (2002) Proc. Natl. Acad. Sci. USA 99, 11405-11410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das A. & Xie, Y. H. (2000) J. Bacteriol. 182, 758-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar R. B., Xie, Y. H. & Das, A. (2000) Mol. Microbiol. 36, 608-617. [DOI] [PubMed] [Google Scholar]
- 20.Smeets L. C. & Kusters, J. G. (2002) Trends Microbiol. 10, 159-162. [DOI] [PubMed] [Google Scholar]
- 21.Bacon D. J., Alm, R. A., Burr, D. H., Hu, L., Kopecko, D. J., Ewing, C. P., Trust, T. J. & Guerry, P. (2000) Infect. Immun. 68, 4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bohne J., Yim, A. & Binns, A. N. (1998) Proc. Natl. Acad. Sci. USA 95, 7057-7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai E. M. & Kado, C. I. (2000) Trends Microbiol. 8, 361-369. [DOI] [PubMed] [Google Scholar]
- 24.Sagulenko V., Sagulenko, E., Jakubowski, S., Spudich, E. & Christie, P. J. (2001) J. Bacteriol. 183, 3642-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt-Eisenlohr H., Domke, N., Angerer, C., Wanner, G., Zambryski, P. C. & Baron, C. (1999) J. Bacteriol. 181, 7485-7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris R. L., Hombs, V. & Silverman, P. M. (2001) Mol. Microbiol. 42, 757-766. [DOI] [PubMed] [Google Scholar]
- 27.Yao X., Jericho, M., Pink, D. & Beveridge, T. (1999) J. Bacteriol. 181, 6865-6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dijkstra A. J. & Keck, W. (1996) J. Bacteriol. 178, 5555-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mushegian A. R., Fullner, K. J., Koonin, E. V. & Nester, E. W. (1996) Proc. Natl. Acad. Sci. USA 93, 7321-7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das A. & Xie, Y. H. (1998) Mol. Microbiol. 27, 405-414. [DOI] [PubMed] [Google Scholar]
- 31.Gomis-Rüth F. X., Moncalián, G., Pérez-Luque, R., González, A., Cabezón, E., de la Cruz, F. & Coll, M. (2001) Nature (London) 409, 637-641. [DOI] [PubMed] [Google Scholar]
- 32.Yeo H. J., Savvides, S. N., Herr, A. B., Lanka, E. & Waksman, G. (2000) Mol. Cell 6, 1461-1472. [DOI] [PubMed] [Google Scholar]
- 33.Krause S., Bárcenz, M., Pansegrau, W., Lurz, R., Carazo, J. M. & Lanka, E. (2000) Proc. Natl. Acad. Sci. USA 97, 3067-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grahn A. M., Haase, J., Bamford, D. H. & Lanka, E. (2000) J. Bacteriol. 182, 1564-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rashkova S., Zhou, X. R., Chen, J. & Christie, P. J. (2000) J. Bacteriol. 182, 4137-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sagulenko E., Sagulenko, V., Chen, J. & Christie, P. J. (2001) J. Bacteriol. 183, 5813-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rashkova S., Spudich, G. M. & Christie, P. J. (1997) J. Bacteriol. 179, 583-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krause S., Pansegrau, W., Lurz, R., de la Cruz, F. & Lanka, E. (2000) J. Bacteriol. 182, 2761-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dang T. A., Zhou, X. R., Graf, B. & Christie, P. J. (1999) Mol. Microbiol. 32, 1239-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones A. L., Shirasu, K. & Kado, C. I. (1994) J. Bacteriol. 176, 5255-5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hapfelmeier S., Domke, N., Zambryski, P. C. & Baron, C. (2000) J. Bacteriol. 182, 4505-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Durfee T., Draper, O., Zupan, J., Conklin, D. S. & Zambryski, P. C. (1999) Yeast 15, 1761-1768. [DOI] [PubMed] [Google Scholar]
- 43.Inoue H., Nojima, H. & Okayama, H. (1990) Gene 96, 23-28. [DOI] [PubMed] [Google Scholar]
- 44.Schiestl R. H. & Gietz, R. D. (1989) Curr. Genet. 16, 339-346. [DOI] [PubMed] [Google Scholar]
- 45.Kuldau G. A., De Vos, G., Owen, J., McCaffrey, G. & Zambryski, P. (1990) Mol. Gen. Genet. 221, 256-266. [DOI] [PubMed] [Google Scholar]
- 46.Baron C., Thorstenson, Y. R. & Zambryski, P. C. (1997) J. Bacteriol. 179, 1211-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vidal M. & Legrain, P. (1999) Nucleic Acids Res. 27, 919-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Das A., Anderson, L. B. & Xie, Y. H. (1997) J. Bacteriol. 179, 3404-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baron C., Llosa, M., Zhou, S. & Zambryski, P. C. (1997) J. Bacteriol. 179, 1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dang T. A. & Christie, P. J. (1997) J. Bacteriol. 179, 453-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anderson L. B., Hertzel, A. V. & Das, A. (1996) Proc. Natl. Acad. Sci. USA 93, 8889-8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spudich G. M., Fernandez, D., Zhou, X. R. & Christie, P. J. (1996) Proc. Natl. Acad. Sci. USA 93, 7512-7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou X. R. & Christie, P. J. (1997) J. Bacteriol. 179, 5835-5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thorstenson Y. R. & Zambryski, P. C. (1994) J. Bacteriol. 176, 1711-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thorstenson Y. R., Kuldau, G. A. & Zambryski, P. C. (1993) J. Bacteriol. 175, 5233-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rambow-Larsen A. A. & Weiss, A. A. (2002) J. Bacteriol. 184, 2863-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bayer M., Iberer, R., Bischof, K., Rassi, E., Stabentheiner, E., Zellnig, G. & Koraimann, G. (2001) J. Bacteriol. 183, 3176-3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Berger B. R. & Christie, P. J. (1994) J. Bacteriol. 176, 3646-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Llosa M., Zupan, J., Baron, C. & Zambryski, P. (2000) J. Bacteriol. 182, 3437-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lai E. M., Chesnokova, O., Banta, L. M. & Kado, C. I. (2000) J. Bacteriol. 182, 3705-3716. [DOI] [PMC free article] [PubMed] [Google Scholar]