Abstract
mRNA degradation provides a powerful means for controlling gene expression during growth, development, and many physiological transitions in plants and other systems. Rates of decay help define the steady state levels to which transcripts accumulate in the cytoplasm and determine the speed with which these levels change in response to the appropriate signals. When fast responses are to be achieved, rapid decay of mRNAs is necessary. Accordingly, genes with unstable transcripts often encode proteins that play important regulatory roles. Although detailed studies have been carried out on individual genes with unstable transcripts, there is limited knowledge regarding their nature and associations from a genomic perspective, or the physiological significance of rapid mRNA turnover in intact organisms. To address these problems, we have applied cDNA microarray analysis to identify and characterize genes with unstable transcripts in Arabidopsis thaliana (AtGUTs). Our studies showed that at least 1% of the 11,521 clones represented on Arabidopsis Functional Genomics Consortium microarrays correspond to transcripts that are rapidly degraded, with estimated half-lives of less than 60 min. AtGUTs encode proteins that are predicted to participate in a broad range of cellular processes, with transcriptional functions being over-represented relative to the whole Arabidopsis genome annotation. Analysis of public microarray expression data for these genes argues that mRNA instability is of high significance during plant responses to mechanical stimulation and is associated with specific genes controlled by the circadian clock.
Keywords: Arabidopsis thaliana, mRNA, stability, circadian, light
Regulation of the stability of mRNAs is an important process in the control of gene expression. This point is perhaps most evident in the wide range of half-lives that is typically observed for nuclear encoded transcripts. In plants, similar to what is reported in mammalian systems, half-lives of mRNAs span several orders of magnitude. Unstable messages have half-lives of less than 60 min and very stable messages on the order of days, with the average being on the order of several hours (1, 2).
Most research has emphasized the study of unstable mRNAs. These transcripts have attracted attention because they often code for regulatory functions that are important for growth and development. For example, mRNAs known to be highly unstable include those for the transcription factors c-myc and c-fos in mammalian cells (3) and the mating-type transcripts in yeast (4). In plants, transcripts that fall into this category include the mRNAs for photo-labile phytochrome (5) and several auxin-inducible transcripts (6, 7). The instability of these mRNAs facilitates fast changes in mRNA levels that result in transient and tightly controlled gene expression (8).
Previous work on unstable transcripts has concentrated on the identification of sequence elements and trans-acting factors that regulate the stability of individual or small groups of transcripts. In eukaryotic cells, transcripts destabilized by multiple overlapping of AUUUA sequences or other AU-rich elements (AREs) located in 3′ untranslated regions (UTRs) have been a major focus (9–12). Several proto-oncogene, cytokine, and transcription factor mRNAs involved in growth and differentiation are recognized for rapid decay via AREs (9–12). In plants, one of the best characterized instability sequences is the DST or downstream element (13, 14). This instability determinant is found in the 3′ UTR of the small auxin up RNA (SAUR) genes. DST elements have a complex structure (15) and the recognition requirements appear to be unique to plants (16). Other sequences that cause instability have also been described (17–19). Nevertheless, the number of structural features identified to date that target transcripts for rapid decay is relatively modest, and many more are likely to be discovered (20).
Although study of individual transcripts is a viable avenue to address this problem, genomic-scale analysis is necessary to evaluate the nature of unstable transcripts within an organism and the regulatory associations they share. Genomic approaches using DNA microarrays have emphasized the study of mRNA levels and how are those levels affected under different conditions (21, 22), but they have been rarely applied to the study of posttranscriptional processes. The first indication that this approach held promise came from data presented in a web site (http://web.wi.mit.edu/young/expression/) referred to in Holstege et al. (23) that estimated stabilities of yeast mRNAs. More recently, Lam et al. (24) used a specialized lymphocyte array to estimate mRNA stabilities in lymphoid cell cultures. Both investigations suggested that global analysis of mRNA stability in intact multicellular organisms should be feasible and more revealing.
In this study, we examined mRNA degradation in intact Arabidopsis plants by using cDNA arrays containing more than 11,000 clones. Similar to the situation in other organisms, the identity and percentage of unstable transcripts in plants had not been evaluated on this scale. Our analysis indicated that at least 1% of the transcripts represented on our arrays decayed with half-lives of less than 60 min. Further, we identified specific functional and regulatory associations among groups of unstable mRNAs that provide insight into the biological significance of rapid mRNA decay mechanisms.
Materials and Methods
Half-Life Measurements and Preparation of RNA Samples.
Half-lives were determined as described by Seeley et al. (5) with the following modifications. Arabidopsis thaliana ecotype Columbia were grown on plates containing 1× Murashige and Skoog salts, 1× Gamborg's vitamins, and 1% sucrose for 2 weeks at 22°C and 16-h/8-h light/dark cycles. The plants were then transferred to a flask with incubation buffer (5). After a 30-min incubation, 3′-deoxyadenosine (cordycepin) was added to a final concentration of 0.6 mM (time 0). Tissue samples were harvested at regular intervals thereafter and quickly frozen in liquid nitrogen. Total RNA was isolated and analyzed by Northern blot using standard techniques. Cordycepin was used to inhibit transcription in these studies because its use is routine in plants (5, 25) in contrast to other inhibitors such as alpha-amanitin, and is more effective in leaf tissue than Actinomycin D (25) presumably because of poor penetration.
Hybridization of cDNA Microarrays.
The 11,521 element cDNA microarray, print name MSU-2_03-00, prepared by the Arabidopsis Functional Genomics Consortium was used in all experiments (26). One hundred micrograms of total RNA corresponding to time 0 and 120 min after cordycepin treatment was labeled during first-strand cDNA synthesis with Cy3- and Cy5-labeled dUTP, respectively, as described (26). Three independent cordycepin treatments (biological replicas) were performed and RNA samples were isolated. Each pair of samples from the 0 and 120 time points was used in two microarray hybridizations, the second with reverse labeling relative to the first (technical replicas). Measurement of the fluorescence corresponding to hybridization intensities was performed with the ScanArray 4000 Microarray Acquisition System (Packard BioChip Technologies, Billerica, MA). We used the SCANALYZE V2.44 software (http://rana.lbl.gov/EisenSoftware.htm) to extract the information of the images generated. The raw data for these experiments is available from the Stanford Microarray Database (SMD; http://genome-www.stanford.edu/microarray/; ref. 27; ExptID: 11374, 11333, 11339, 11323, 11375, and 11342).
Microarray Data Analysis.
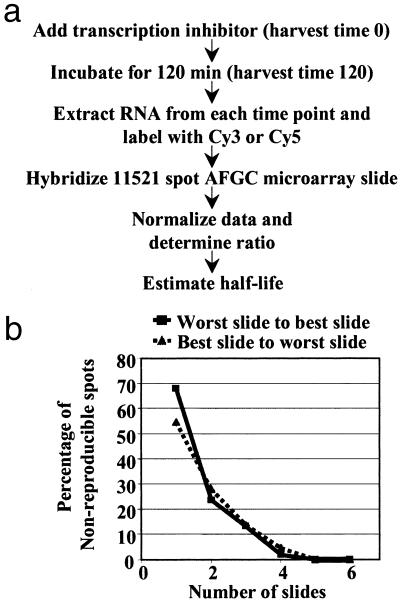
Stringent quality control measures were applied to define the working data set. Spots with abnormal shapes or high local background were discarded manually. Spots with channel intensity values smaller than the mean plus two standard deviations of each slide background or with GTB2 values smaller than 0.65 in more than two channels were discarded because of low signal. The quality of the hybridization was also evaluated by visual inspection of the gradients, using the most sensitive setting of the “Array Color Plot” tool implemented in SMD. Slides that showed gradients in more than 25% of the array surface and/or that had R2 values >0.15 (indicating a strong dependence on spatial location) were not used for data analysis. The percent of the array surface that exhibited gradients was also used to order the slides from worst to best or best to worst in Fig. 1.
Fig 1.
Strategy for monitoring mRNA stability using cDNA microarrays. (a) RNA samples corresponding to 0 and 120 min after the addition of the transcriptional inhibitor cordycepin were labeled with Cy3 and Cy5, respectively, and used to hybridize 11K microarray slides. Each pair of RNA samples was reverse labeled for a separate microarray hybridization. These hybridizations were performed with samples from three independent cordycepin treatments for a final data set of six slides. Half-life values were then estimated from the normalized ratios. (b) Nonreproducible spots decrease as a function of the number of slides, nearly leveling out when the data from four slides is combined. The quality of the slides, best to worst or worst to best based on the extent of visible gradients (see Materials and Methods), does not significantly affect the reproducibility of the data when two or more slides are considered, although the curves are slightly steeper with better slides.
The Z-score method in log space with a 90% trimmed data set was used for global normalization of the data (26). The difference in mRNA levels between the time points considered (0 and 120 min) can be used to estimate the rates of decay by using the equation ln(Normalized Ratio) = −kdecayt, with the half-life being t1/2 = 0.693/kdecay, because mRNA degradation generally obeys first-order kinetics (24). Statistical analysis of the ratios was performed using the t test as described in the text.
Sequence and Gene Expression Analysis.
Sequences of 59 clones representing Arabidopsis thaliana genes for unstable transcripts (AtGUTs) were determined and found to be consistent with sequences deposited in GenBank. For expressed sequence tag (EST) identification, the BLASTN program was used to search the completed Arabidopsis genome sequence downloaded from The Institute for Genomic Research (TIGR). Functional categories were obtained from the Munich Information Center for Protein Sequences. For analysis of gene expression data across multiple experiments, the CLUSTER and TREEVIEW software were used (ref. 28; http://rana.lbl.gov/EisenSoftware.htm). Visual images were generated with the TREEVIEW software by using the output generated by the hierarchical clustering program of the CLUSTER software. The uncentered correlation similarity metric was used to perform average linkage clustering.
Sequences of AtGUTs (5′ UTR, coding sequence, 3′ UTR) were obtained from the TIGR Arabidopsis genome as described in the supporting information, which is published on the PNAS web site, www.pnas.org. Frequencies of overlapping (1-bp window) oligonucleotides (up to 6 nt in length) were determined in the 3′ UTR sequences of AtGUTs and also in the 3′ UTR sequences of a control set of genes as described by van Helden et al. (29). The control set was derived from 4,064 clones corresponding to stable transcripts on the array. To assess significance, 1,000 random samples of the same size as the test set were taken from the control sequences and oligonucleotide frequencies were determined in these samples. The criteria for significance were as follow: (i) oligonucleotide was at least 2-fold more abundant in unstable than in whole control set; (ii) oligonucleotide had frequencies >2 SD above mean frequency in the 1,000 random samples from control set; (iii) oligonucleotide was present in >10% of test sequences. Additional MEME searches (30) were carried out as described at http://meme.sdsc.edu. Various combinations of the MEME parameters were tested: motif distribution, 1–3; number of motifs, 3–5; motif width, 5–50. Programs written in the Practical Extraction and Report Language (Perl) were used for sequence extraction and manipulation.
Results and Analysis
Monitoring mRNA Stability by Using cDNA Microarrays.
mRNA decay rates, expressed as half-life values, are typically measured by monitoring the disappearance of a transcript by Northern blot after transcription of the corresponding gene has been halted. We combined this simple experimental strategy with the highly parallel power of DNA microarray analysis (31) as outlined in Fig. 1a. Total RNA samples corresponding to 0- and 120-min time points after transcriptional inhibition with cordycepin were isolated. One hundred micrograms of total RNA from each of these samples was used to synthesize cDNA probes by incorporating Cy3- or Cy5-labeled dUTP during oligo(dT)-primed reverse transcription. The probes were combined and used for hybridization of the 11,521-elements cDNA microarray (11K microarray) prepared on glass slides by the Arabidopsis Functional Genomics Consortium. We performed three biological replica experiments, each with a reverse-labeling technical replicate. The purpose of these repetitions was to increase the likelihood of detecting significant differences in mRNA levels, while decreasing the likelihood of false positives, which might be common on microarray studies with one or two slides (32). Quite reasonably, the number of nonreproducible normalized intensity ratios ≥2 decreased as a function of the number of slides, nearly leveling out below 5% when four slides were considered (Fig. 1b). Based on this data, and to be rigorous, we defined our working data set as all those clones with reproducible normalized intensity ratios ≥2 in five of six slides.
At Least 1% of Clones on the 11K Arabidopsis Microarrays Correspond to Unstable Messages.
To identify and characterize the most unstable transcripts from our working data set, we focused our attention on the transcripts that were most diminished after treatment with cordycepin for 120 min. Clones whose median normalized intensity ratios were ≥4 (0 vs. 120 min) and that met several quality control criteria (see Materials and Methods) were used for further analysis. In this study, ESTs that overlap with the same annotated ORF were considered as representing the same gene. This is a reasonable assumption because the expression patterns of groups of ESTs that match the same ORF were well correlated across multiple experiments (data not shown). Based on this criterion, the selected clones corresponded to 100 genes that were termed Arabidopsis thaliana genes with unstable transcripts (AtGUTs), because the calculated half-lives of the encoded transcripts were 60 min or less (see Table 2, which is published as supporting information on the PNAS web site). Similarly, we identified 225 genes with moderately unstable mRNAs, whose estimated half-lives ranged from 60 to 120 min. The great majority of the transcripts in Arabidopsis appeared to decay with rates greater than 2 h, consistent with the idea that most messages in plants are relatively stable (1).
The 11K microarray used in these studies represents an estimated 7,800 unique genes (26) so the 100 AtGUTs we identified correspond to about 1%. This number likely represents an underestimate of unstable Arabidopsis transcripts, especially when extrapolated to the whole genome for several reasons. First, unstable mRNAs are often associated with low steady state levels, which may be underrepresented in the EST collections used for the 11K microarray. Second, some unstable transcripts likely fall below the detection limits of the microarray technique or might not meet our stringent reproducibility requirements. However, the channel intensity distribution for AtGUTs resembled that of the whole array, suggesting the AtGUTs identified were not strongly biased to either high or low expression levels on the 11K microarray. The identification of highly expressed AtGUTs was an added bonus from this analysis because these transcripts should greatly facilitate future studies of steps in their degradation. Finally, multiple members of closely related gene families may be missed because the 11K microarray was not designed to resolve gene family members. Thus, on the basis of our work, it seems valid to estimate that at least 1% of the genes of Arabidopsis correspond to unstable transcripts.
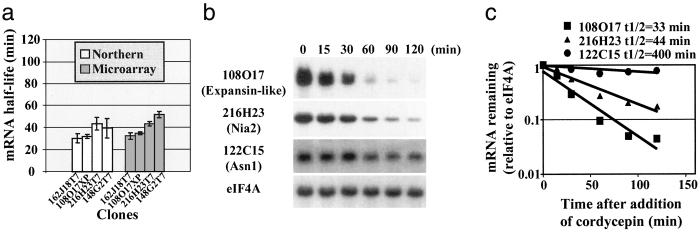
We used conventional Northern blot analysis of cordycepin time courses with several time points to confirm the 11K microarray data. Four randomly selected transcripts with half-lives of less than 60 min showed comparable turnover rates in full cordycepin time courses (Fig. 2a). Two representative examples are shown in Fig. 2 b and c. In addition, we assessed the statistical significance of the ratio values for the genes of interest. When using the t test and the conservative Bonferroni method to adjust P values (33), all selected AtGUTs showed significantly different ratios from the mean of the population at α < 0.0001 (see Table 2). Several genes with moderately unstable messages according to the microarray studies were at least moderately unstable in Northern blots (data not shown). Four stable transcripts according to microarray data were also found stable by Northern blot analysis further validating our results. Two representative examples of these stable transcripts are shown in Fig. 2 b and c.
Fig 2.
Confirmation of the instability of transcripts identified by microarray analysis. (a) Half-life values determined by Northern blot are comparable to estimates from microarray analysis for four randomly selected unstable transcripts. Northern blot values are representative of at least three independent cordycepin time courses. (b) Representative Northern blot analysis of cordycepin time courses for two randomly selected unstable and two stable transcripts. Samples consisted of 10 μg of total RNA isolated from the indicated time points. (c) Quantitation of the decrease in mRNA abundance and half-life estimation. The signal for eIF-4A does not change significantly during the time courses and was used as a reference for equal loading.
General Structural Features of Genes with Unstable and Stable Transcripts Are Similar.
The identification of the AtGUTs allowed us to evaluate them for structural properties that might play a role in determining their instability. We compared the sequences of the 100 AtGUTs against genes that encode stable transcripts under our conditions. AtGUTs were evenly distributed throughout the Arabidopsis genome and showed no significant differences in nucleotide composition, number of introns, size of the coding sequence, and codon usage as compared with genes with stable mRNAs.
We did not expect to find a simple sequence that would be present in the 3′ UTR of all or most AtGUTs because previous observations suggest that many instability sequences exist (17–19). Consistent with this prediction, neither an oligonucleotide frequency approach (29) nor a probabilistic approach using the MEME software (30) was indicative of a simple sequence common to all or most AtGUTs compared with controls. However a few AtGUTs have potential AU-rich element (ARE)-like instability sequences (3, 10, 11), typified by repeats of the AUUUA motif: a putative nematode-resistance gene (At2g4000) that encodes the most unstable transcript in our conditions and two genes of unknown function (At1g72450 and At2g41640). Although the functional significance of these sequences remains to be determined, these transcripts are potential targets for the AUUUA-mediated decay pathway in Arabidopsis. Similarly, two AtGUTs, the senescence-associated gene sen1 (At4g35770) and a putative light-regulated gene similar to the ccr gene from Citrus paradisi (At3g26740), have DST-like elements in their 3′ UTRs. Interestingly, the expression of these two transcripts is altered in dst1, a mutant deficient in DST-mediated decay (34). Therefore, these transcripts are potential primary targets of the DST-mediated decay pathway in Arabidopsis (34).
AtGUTs Are Predicted to Play a Role in a Broad Range of Cellular Processes but Most Prominently in Transcription.
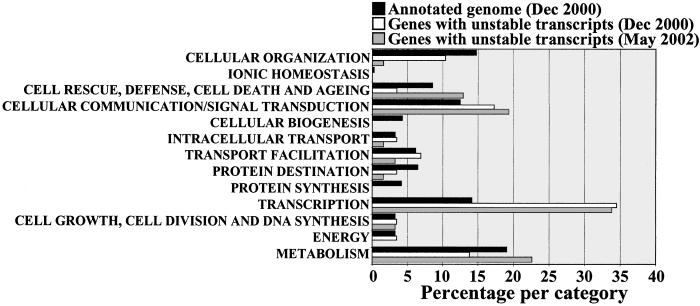
To explore the potential cellular roles of AtGUTs, we analyzed how they were distributed among the functional categories assigned by the Munich Information Center for Protein Sequences (Fig. 3). More than half of the AtGUTs could be assigned to a Munich Information Center for Protein Sequences category (see Table 2), with the remainder lacking homology to known proteins. The distribution of predicted functions for AtGUTs suggests that they participate in a broad range of plant processes and in roughly the same proportion as the whole complement of Arabidopsis genes. Interestingly, enrichment was observed for transcriptional functions. AtGUTs encode transcriptional functions more than twice the expected frequency based on the whole Arabidopsis genome annotation (35). BLAST search analysis (36) indicated that 14 of the 21 AtGUTs that belong to this transcriptional class were not found in the sequenced genome of Homo sapiens, Mus musculus, Rattus norvegicus, Caenorhabditis elegans, Drosophila melanogaster, Saccharomyces cerevisiae, Synechocystis, Eubacteria, and Archebacteria (BLASTX program, P < 0.01; Table 1). This is in line with the detailed analysis of Arabidopsis transcription factors performed by Riechmann et al. (37), which indicated that 45% of those annotated on the genome are from families specific to plants, reflecting the independent evolution of many plant transcription factors. It is possible that plants might also have evolved mechanisms that are distinct from those of other eukaryotes for regulating the stability of these transcripts. Plant-specific mechanisms might not be exclusive to transcriptional functions but could also extend to other AtGUTs, which are unique to plants.
Fig 3.
Instability is associated with a broad range of plant processes. Genes with unstable transcripts were classified according to the scheme of Munich Information Center for Protein Sequences. AtGUTs are predicted to participate in a broad range of cellular processes with transcriptional functions over-represented compared with what is expected based on the whole genome annotation. To allow comparison, AtGUTs were classified based on the information released for the annotation of the whole A. thaliana genome sequence in December 2000. The most updated annotation for the AtGUTs is also included (May 2002), although a whole genome annotation based on this updated information is not yet available.
Table 1.
Arabidopsis genes with unstable messages that belong to the Munich Information Center for Protein Sequences transcriptional category (04) as of May 2002
| Locus | Description |
|---|---|
| At1g13260 | DNA-binding protein RAV1 |
| At1g19180 | Hypothetical protein |
| At1g32640 | Putative protein kinase; bHLH protein |
| At2g22430 | Homeodomain transcription factor (ATHB-6) |
| At2g24570 | Putative WRKY-type DNA binding protein |
| At2g40140 | Putative CCCH-type zinc finger protein |
| At3g15210 | Ethylene responsive element binding factor 4 |
| At3g16720 | Putative RING zinc finger protein |
| At3g44260 | CCR4-associated factor 1-like protein |
| At3g54810 | Similar to GATA transcription factor 3 |
| At4g12040 | Similar to zinc finger protein ZNF216-M. musculus |
| At4g17230 | Scarecrow-like 13 (SCL13) |
| At4g17500 | Ethylene responsive element binding factor 1-like |
| At4g31550 | WRKY family transcription factor |
| At5g04340 | Putative c2h2 zinc finger transcription factor |
| At5g05440 | Putative protein |
| At5g07580 | Transcription factor-like protein |
| At5g59550 | Similar to COP1-interacting protein CIP8 |
| At5g61590 | Ethylene responsive element binding factor 5-like |
| At5g61600 | Ethylene responsive element binding factor 4-like |
| At5g63790 | Putative protein |
It should be noted that this category includes transcription as well as other aspects of RNA metabolism.
Plant-specific genes as determined by blast search analysis (June 2000).
Plant-specific genes according to the classification by Reichman et al. (37).
Rapid mRNA Degradation Is Associated with Arabidopsis Responses to Mechanical Stimulation and Circadian Rhythms.
To begin understanding the physiological implications of instability in Arabidopsis, we examined expression of the identified AtGUTs for patterns of regulation. Hierarchical cluster analysis was performed (see Materials and Methods) using the publicly available microarray data for the AtGUTs, deposited in the Stanford Microarray Database (27) by the Arabidopsis Functional Genomics Consortium. At the time of this study, 112 slides corresponding to 47 different experiments carried out under various treatments, environmental conditions, or developmental stages, or in different genotypes, were available.
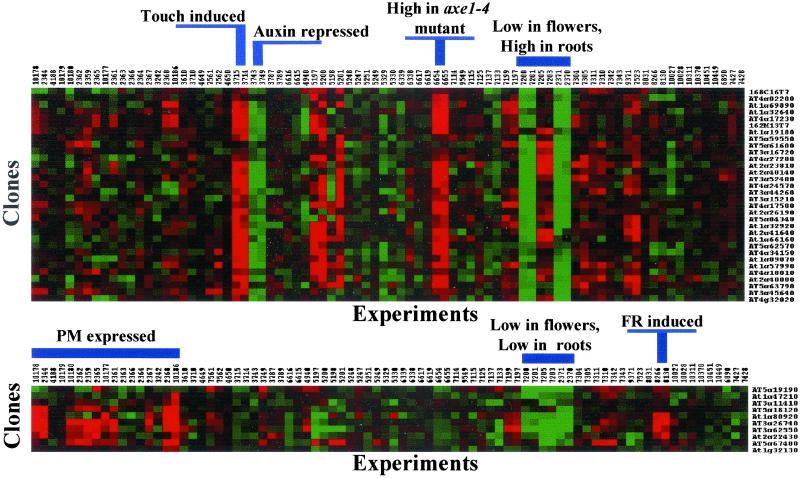
Two main clusters of genes were observed. The largest contained 32 genes, the majority of which were induced by mechanical stimulation (touch; see Table 3, which is published as supporting information on the PNAS web site, for SMD experiment identifiers; Fig. 4). Several of the genes in this cluster also appeared repressed in an auxin treatment and induced in the histone deacetylase mutant axe1–4 relative to wild-type Arabidopsis plants (38). In addition, most showed organ-preferential expression with low levels in flowers and high levels in roots compared with a reference sample prepared from a mixture of plant organs (Fig. 4, and see Table 3). The identification of a touch-induced cluster of AtGUTs is consistent with touch responses being fast (39), and instability being critical when rapid changes in mRNA steady state levels are to be achieved. In fact, the touch gene transcripts initially characterized were detected within minutes of treatment and disappeared very rapidly thereafter, consistent with rapid turnover (39). Interestingly, 32 (34%) of the 95 genes induced by the touch treatment in SMD encode unstable transcripts (see Table 3). In contrast, only 0.8% of the genes repressed under high CO2 conditions (experiments 7561 and 7562) and 5% of the genes induced after 1.5 and 3 h H2O2/NO2 cotreatment (experiments 7523 and 9371) did the same. The number of genes differentially expressed in the CO2 and H2O2/NO2 experiments is in the same range as the overall number of AtGUTs identified. These results indicate that not all physiological responses have an equal instability component. Moreover, our data suggest that mRNA instability is an important regulatory component of the touch response.
Fig 4.
Cluster analysis indicates that a set of AtGUTs is induced by mechanical stimulation (touch) and another is controlled in a diurnal fashion. Hierarchical cluster analysis of expression data for the AtGUTs across multiple microarray experiments was performed with cluster and treeview software (28). Expression characteristics shared by genes in each cluster are indicated (PM, post meridian; FR, far-red light). Each row represents a gene and each column represents an experiment. Small labels on top of the clusters are SMD experiment identifiers. Small labels on right side of clusters indicate A. thaliana loci or EST clone identifiers.
It is relevant to note that we were unable to confirm the auxin regulation shown by genes inside the touch cluster. Indole acetic acid (IAA) treatment of 2-week-old Arabidopsis plants did not affect the expression of three different genes in the touch cluster over a 24-h time course (data not shown). These three genes showed a transient induction irrespective of the presence or absence of IAA in the solution sprayed. However, IAA treatment strongly induced the expression of the small auxin up RNA gene SAUR-AC1 (6) compared with the control (data not shown). Our data suggest that, at least in the conditions tested, the genes that belong to the touch cluster are not regulated by the phytohormone auxin, but are induced by spraying alone. Presumably the genes in the touch cluster are more sensitive to experimental variation, especially if there is mechanical stimulation involved.
A second smaller cluster contained six genes whose expression was regulated diurnally (Fig. 4). In addition, some of these genes were induced by a far-red light treatment and showed organ-preferential expression with low levels in flowers and roots (Fig. 4). Interestingly, some of these genes (At5g67480, At3g62550, At3g26740, and At1g80920) have been also shown to be controlled by the circadian clock with a peak in mRNA abundance in the afternoon (26). Further, 12 other AtGUTs (At3g15450, At1g37130, At1g75900, At4g31500, At2g39730, At2g32150, At1g13260, At3g55240, At2g35260, At1g49500, At2g29450, and At4g32060) have been reported as clock-controlled genes (see Table 3; refs. 26 and 40). At similar transcriptional rates, different mRNA stabilities would translate into circadian mRNA profiles with distinct phases and amplitudes (41). Therefore, instability might be essential for the mRNA oscillatory patterns observed for specific AtGUTs that are regulated by the clock. Posttranscriptional regulation of mRNA stability has been shown to play a role in the expression of the Drosophila clock gene per (41). There is also evidence that transcription makes a small contribution to the observed circadian expression pattern of the Arabidopsis NIA2 gene (42). Hence, modulation of mRNA stability could also contribute to the clock-regulated expression of specific AtGUTs. An interesting common feature of the genes in the touch and diurnal cluster was their low expression level in flowers (Fig. 4). In fact, 40% of the AtGUTs identified showed similar diminished expression suggesting rapid mRNA turnover might not be as prominent in flowers as in other organs. Perhaps reproductive tissues do not require rapid response control as much as vegetative tissues do, but instead favor the more economical long-lived mRNAs that are common in specialized cells (43).
Because genes that showed similar patterns of expression might share regulatory mechanisms, the 3′ UTR sequences of AtGUTs that belong to the touch and light regulated clusters were analyzed for the presence of common sequence elements as described previously. Neither the oligonucleotide frequency approach nor the MEME software provided candidate sequence motifs overrepresented among these subsets of AtGUTs. Signals that control mRNA metabolism are often composite sequence elements—e.g., mRNA localization signals (44)—whose key motifs have been difficult to identify. Alternatively, a sequence conserved in a small number of AtGUTs might not be detected by our approach.
Perhaps the most intriguing question for future studies prompted by our work relates to the AtGUTs regulated by touch and light. Are these AtGUTs “constitutively” unstable, or is their mRNA stability regulated in response to light, touch, or other signals? The approach we describe has the potential to address this and other exciting questions about posttranscriptional processes in plants. By comparing global turnover rates under different conditions, posttranscriptional regulatory networks might be unraveled. This large-scale approach for analysis of posttranscriptional control mechanisms should help uncover the extent and significance of this level of regulation in plants in response to a variety of stimuli.
Supplementary Material
Acknowledgments
We thank Dr. Robert Schaffer, Dr. Jeff Landgraf, Dr. Ellen Wisman, and the Green lab members for helpful discussions, the other members of the Arabidopsis Functional Genomics Consortium for technical advice, Dr. Ambro van Hoof for critical comments on the manuscript, and Dr. Tom Newman for providing and sequencing the EST clones. This work was funded by U.S. Department of Agriculture Grant 2000-01491, Department of Energy Grant DE-FG02-91ER20021, and National Science Foundation Grant DBN987638 (to P.J.G.).
Abbreviations
11K microarray, 11,521-element cDNA microarray prepared on glass slides by the Arabidopsis Functional Genomics Consortium
SMD, Stanford Microarray Database
UTR, untranslated region
EST, expressed sequence tag
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Siflow C. D. & Key, J. L. (1979) Biochemistry 18, 1013-1018. [DOI] [PubMed] [Google Scholar]
- 2.Hargrove J. L., Hulsey, M. G. & Beale, E. G. (1991) BioEssays 13, 667-674. [DOI] [PubMed] [Google Scholar]
- 3.Greenberg M. E. & Belasco, J. G. (1993) in Control of Messenger RNA Stability, eds. Belasco, J. G. & Brawerman, G. (Academic, San Diego), pp. 199–218.
- 4.Peltz S. W. & Jacobson, A. (1992) Curr. Opin. Cell Biol. 4, 979-983. [DOI] [PubMed] [Google Scholar]
- 5.Seeley K. A., Byrne, D. H. & Colbert, J. T. (1992) Plant Cell 4, 29-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McClure B. A. & Guilfoyle, T. J. (1989) Science 243, 91-93. [DOI] [PubMed] [Google Scholar]
- 7.Koshiba T., Ballas, N., Wong, L. M. & Theologis, A. (1995) J. Mol. Biol. 253, 396-413. [DOI] [PubMed] [Google Scholar]
- 8.Treisman R. (1985) Cell 42, 889-902. [DOI] [PubMed] [Google Scholar]
- 9.Shaw G. & Kamen, R. (1986) Cell 46, 659-667. [DOI] [PubMed] [Google Scholar]
- 10.Ohme-Takagi M., Taylor, C. B., Newman, T. C. & Green, P. J. (1993) Proc. Natl. Acad. Sci. USA 90, 11811-11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen C.-Y. A. & Shyu, A.-B. (1995) Trends Biochem. Sci. 20, 465-470. [DOI] [PubMed] [Google Scholar]
- 12.Vasudevan S. & Peltz, S. W. (2001) Mol. Cell 7, 1191-1200. [DOI] [PubMed] [Google Scholar]
- 13.McClure B. A., Hagen, G., Brown, C. S., Gee, M. A. & Guilfoyle, T. J. (1989) Plant Cell 1, 229-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newman T. C., Ohme-Takagi, M., Taylor, C. B. & Green, P. J. (1993) Plant Cell 5, 701-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sullivan M. L. & Green, P. J. (1996) RNA 2, 308-315. [PMC free article] [PubMed] [Google Scholar]
- 16.Feldbrügge M., Aritzi, P., Sullivan, M. L., Zamore, P. D., Belasco, J. G. & Green, P. J. (2002) Plant Mol. Biol. 49, 215-223. [DOI] [PubMed] [Google Scholar]
- 17.Ross J. (1995) Microbiol. Rev. 59, 423-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caponigro G. & Parker, R. (1996) Microbiol. Rev. 60, 233-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gutiérrez R. A., MacIntosh, G. C. & Green, P. J. (1999) Trends Plant Sci. 4, 429-438. [DOI] [PubMed] [Google Scholar]
- 20.Taylor C. B. & Green, P. J. (1995) Plant Mol. Biol. 28, 27-38. [DOI] [PubMed] [Google Scholar]
- 21.Brown P. O. & Botstein, D. (1999) Nat. Genet. Suppl. 21, 33-37. [DOI] [PubMed] [Google Scholar]
- 22.Schaffer R., Landgraf, J., Perez-Amador, M. & Wisman, E. (2000) Curr. Opin. Plant Biol. 11, 162-167. [DOI] [PubMed] [Google Scholar]
- 23.Holstege F. C. P., Jennings, E. G., Wyrick, J. J., Lee, T., Hengartner, C. J., Green, M. R., Golub, T. R., Lander, E. S. & Young, R. A. (1998) Cell 95, 717-728. [DOI] [PubMed] [Google Scholar]
- 24.Lam L. T., Pickeral, O. K., Peng, A. C., Rosenwald, A., Hurt, E. M., Giltnane, J. M., Averett, L. M., Zhao, H., Davis, R. E., Sathyamoorthy, M., et al. (2001) Genome Biol. 2, 1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson M. A., Pérez-Amador, M. A., Lidder, P. & Green, P. J. (2000) Proc. Natl. Acad. Sci. USA 97, 13991-13996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaffer R., Landgraf, J., Accerbi, M., Simon, V., Larson, M. & Wisman, E. (2001) Plant Cell 13, 113-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherlock G., Hernandez-Boussard, T., Kasarskis, A., Binkley, G., Matese, J. C., Dwight, S. S., Kaloper, M., Weng, S., Jin, H., Ball, C. A., et al. (2001) Nucleic Acids Res. 29, 152-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eisen M. B., Spellman, P. T., Brown, P. O. & Bolstein, D. (1998) Proc. Natl. Acad. Sci. USA 95, 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Helden J., André, B. & Collado-Vides, J. (1998) J. Mol. Biol. 281, 827-842. [DOI] [PubMed] [Google Scholar]
- 30.Bailey T. L. & Elkan, C. (1994) in Proceedings on the Second International Conference on Intelligent Systems for Molecular Biology, eds. Altman, R., Brutlag, D., Karp, P., Lathrop, R. & Searls, D. (AAAI, Menlo Park, CA), pp. 28–36.
- 31.Schena M., Shalon, D., Davis, R. W. & Brown, P. O. (1995) Science 270, 467-470. [DOI] [PubMed] [Google Scholar]
- 32.Ting Lee M.-L., Kuo, F. C., Whitmore, G. A. & Sklar, J. (2000) Proc. Natl. Acad. Sci. USA 97, 9834-9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samuels M. L. (1989) in Statistics for the Life Sciences, ed. Samuels, M. L. (Dellen, San Francisco), pp. 504–505.
- 34.Pérez-Amador M. A., Lidder, P., Johnson, M. A., Landgraf, J., Wisman, E. & Green, P. J. (2002) Plant Cell 13, 2703-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.The Arabidopsis Genome Initiative (2000) Nature (London) 408, 796-815. [DOI] [PubMed] [Google Scholar]
- 36.Altschul S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403-410. [DOI] [PubMed] [Google Scholar]
- 37.Riechmann J. L., Heard, J., Martin, G., Reuber, L., Jiang, C.-Z., Keddie, J., Adam, L., Pineda, O., Ratcliffe, O. J., Samaha, R. R., et al. (2000) Science 290, 2105-2110. [DOI] [PubMed] [Google Scholar]
- 38.Murfett J., Wang, X.-J., Hagen, G. & Guilfoyle, T. J. (2001) Plant Cell 13, 1047-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Braam J. & Davis, R. W. (1990) Cell 60, 357-364. [DOI] [PubMed] [Google Scholar]
- 40.Harmer S. L., Hogenesch, J. B., Straume, M., Chang, H.-S., Han, B., Zhu, T., Wang, X., Kreps, J. A. & Kay, S. A. (2000) Science 290, 2110-2113. [DOI] [PubMed] [Google Scholar]
- 41.So W. V. & Rosbash, M. (1997) EMBO J. 16, 7146-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pilgrim M. L., Caspar, T., Quail, P. H. & McClung, C. R. (1993) Plant Mol. Biol. 23, 349-364. [DOI] [PubMed] [Google Scholar]
- 43.Weiss I. M. & Liebhaber, S. A. (1994) Mol. Cell. Biol. 14, 8123-8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bashirullah A., Cooperstock, R. L. & Lipshitz, H. D. (1998) Annu. Rev. Biochem. 67, 335-394. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.