Abstract
Background
Hypohidrotic ectodermal dysplasia (HED) is a rare genetic disorder that affects the development of the skin, hair, nails, teeth, and sweat glands. Due to its rarity, there are currently few methods that can be applied to facilitate its diagnosis during the prenatal period. Although a prenatal ultrasonographic examination will detect early signs of the disease, there are few reports on specific prenatal ultrasonographic features of ectodermal dysplasia. Genetic diagnosis can confirm the disease, but the numerous gene variants that cause ectodermal dysplasia have not been fully identified.
Case presentation
Our case was a multiparous woman carrying a single male fetus who underwent a fetal ultrasound examination at 23 weeks of gestation. The examination revealed thin alveolar bone, with no presence of hypoechoic tooth germ tissue in both the upper and lower alveolar bones. The seven-year-old male proband in this family manifested a clinical phenotype of sparse hair and underdeveloped teeth, and trio-based whole-exome sequencing (WES) performed on both parents and the proband revealed a novel and likely pathogenic variant of the EDA gene (NM_001399.4: c.806G > T, p.Gly269Val) associated with X-linked HED (XLHED; OMIM:305100). Based on the results of the fetal ultrasound examination and the results of the proband’s genetic testing, the couple ultimately decided to terminate the pregnancy. The DNA of the fetal skin tissue after the induced abortion was extracted for Sanger sequencing, and it was confirmed that the fetus possessed ectodermal dysplasia generated by the afore-mentioned EDA gene mutation.
Conclusions
Our study suggested that prenatal ultrasonography constituted an effective method for screening ectodermal dysplasia during pregnancy. In addition, our findings expanded the range of EDA variants in XLHED patients; and this discovery may now assist potential patients in receiving an accurate diagnosis, allowing them to make appropriate reproductive decisions.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12884-025-07963-9.
Keywords: Hypohidrotic ectodermal dysplasia, Prenatal ultrasound screening, Whole-exome sequencing, EDA c.806G > T
Background
Ectodermal dysplasia (ED) is a rare genetic disorder that primarily affects tissues and organs of ectodermal origin, and it is classified as either hidrotic or hypohidrotic depending upon whether the sweat glands are involved. Patients with hypohidrotic ectodermal dysplasia (HED) usually exhibit a series of typical clinical features. Follicular hypoplasia causes patients to show symptoms such as missing eyebrows and eyelashes and sparse, fine, and lightly pigmented scalp hair; while dental dysplasia is characterized by hypodontia and peg-shaped teeth. Dysfunction of sweat glands results in an imbalance in body temperature regulation that can then lead to hyperthermia during physical activity or exposure to a warm environment. In addition, patients with HED exhibit a distinctive facial appearance that is characterized by a prominent forehead; saddle nose; and full, everted lips [1, 2]. Hypohidrotic ectodermal dysplasia encompasses a variety of inheritance patterns that principally include X-linked recessive inheritance, autosomal dominant inheritance, and autosomal recessive inheritance—each of which is closely related to specific gene anomalies. X-linked recessive inheritance is the most common inheritance pattern for HED, and it primarily involves the EDA (ectodysplasin A) gene located on the X chromosome. Abnormal function of the ectodysplasin A protein encoded by the EDA gene disrupts the normal functioning of the EDA-EDAR-EDARADD-signaling pathway, modulating the expression of downstream genes and cellular behavior; and this perturbation ultimately leads to abnormal development of ectodermal tissues such as hair, teeth, sweat glands, skin, and nails [3, 4]. Autosomal dominant and recessive inheritance patterns are relatively rare in HED, and investigators have reported that variants in the DLX gene family can precipitate autosomal dominant or recessive HED [5]. Certain variants in the DLX3 gene (OMIM: 600525) alter the gene’s ability to bind to DNA and are partially responsible for dental deformities and changes in facial features, including a prominent forehead and a collapsed nose bridge [6, 7]. Variants in the WNT10A gene (OMIM: 606268) induce autosomal recessive ED by altering the WNT-signaling pathway [8]. We herein reported X-linked hypohidrotic ectodermal dysplasia (XLHED; OMIM:305100) due to an EDA gene (OMIM:300451) variant.
Prenatal diagnosis is crucial for XLHED, and its early diagnosis can provide parents with information on the condition of their fetus and assist them in rendering reasonable fertility decisions. Such a timely diagnosis can also provide a basis for the perinatal management and early postnatal intervention in confirmed fetuses, reducing the adverse effects of the disease on neonates. Pigno et al. [9] showed that XLHED was strongly associated with tooth dysplasia; Hammersen J et al. [10] proposed that ultrasonographic examination of dental germs was highly specific and reliable in prenatal screening for XLHED; and Li et al. [11] reported prenatal ultrasonographic findings in HED patients over the past 20 years. Most of the latter group’s patients (11/13) exhibited no tooth germ tissue or reduced numbers of tooth germs, while others (2/13) also showed mandibular hypoplasia and thick and everted upper and lower lips. We reported the same ultrasonographic features, further confirming the reliability of these features in the prenatal diagnosis of XLHED.
If the fetus is found to manifest some characteristic ultrasound manifestations related to HED during routine prenatal ultrasonographic screening, further genetic testing should be performed so as to determine whether the fetus possesses HED and to discern the specific type. Among the many gene mutations that cause the disease, variants in the EDA gene are the most common, accounting for over half of all cases [12]. However, the genetic variants that generate ED have not yet been fully elucidated. We herein identified a novel EDA variant through whole-exome sequencing (WES) and reviewed all the variants in exon 7, where the variant was located, and determined which variants were classified as pathogenic or likely pathogenic in the ClinVar database. This methodology allowed us to better understand the position of the novel EDA variant in the entire exon 7 variation spectrum and to explore the possible underlying pathogenic mechanism(s) involved.
Case presentation
A 34-year-old multiparous woman with a history of three pregnancies and two deliveries was referred to the authors’ Department for genetic counseling during the 23rd week of her current pregnancy. Her first child was a healthy female, while her second child (the proband) presented as a seven-year-old boy who exhibited primary clinical symptoms of hypotrichosis and dental agenesis, with only one upper molar (Fig. 1). During this pregnancy, an ultrasonogram at 12 + 2 weeks of gestation showed a nuchal translucency (NT) measurement of 2.0 mm. Given the significant family history, the woman was thus referred to the authors’ Department for genetic counseling, and a systematic ultrasonographic examination conducted at the authors’ Department revealed thin upper and lower alveolar ridges with no evident tooth germ tissue in the inner alveolar region. We additionally observed an abnormally pointed arc of the lower alveolar bone and eversion of both the upper and lower lips (Fig. 2). After a detailed consultation, the pregnant woman decided to terminate the pregnancy after amniocentesis, and autopsy results after the induction of labor confirmed the ultrasound findings (Supplementary Materials). Peripheral blood was also drawn from the proband and the couple for WES.
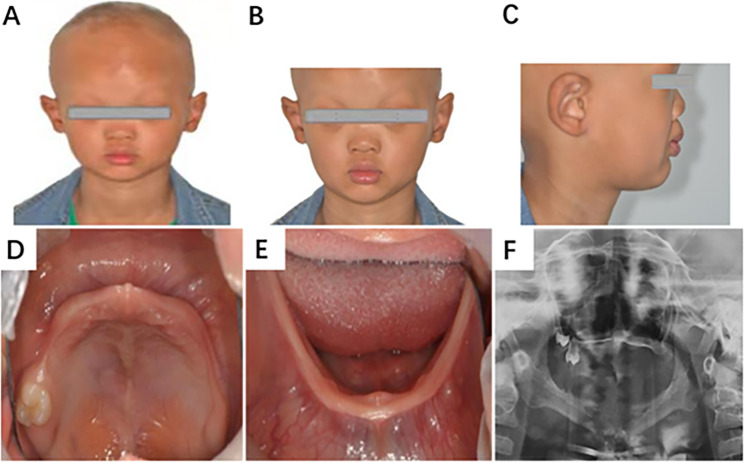
Fig. 1.
Clinical features of patients with an EDA variation. A The proband shows dry and rough skin, gray complexion, thinning and lusterless hair, and partial loss of hair on the top occipital region of the head. B Thinning, raised, and light eyebrows. C A depressed nasal bridge and saddle nose. D Peg-like teeth in the upper alveolar bones. E A missing tooth in the lower alveolar bones. F Panoramic radiograph of the proband
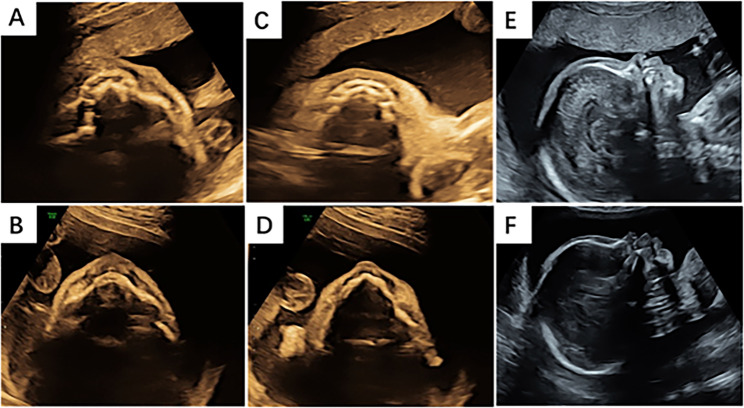
Fig. 2.
Comparison of facial ultrasound features between a normal fetus and a fetus with HED. A A healthy control fetus of the same gestational age. The axial-view image portrays normal alveolar bone as represented by round hypoechoic tooth germs that are arranged in an arch-like shape in the upper alveolar bone. B The fetus in our case. The axial-view image of the upper alveolar bone shows no hypoechoic tooth germs. C The axial-view image shows normal alveolar bone as represented by round hypoechoic tooth germs that are arranged in an arch-like shape in the upper alveolar bone. D The fetus in our case. The axial view shows the sharpened lower alveolar bone and no hypoechoic tooth germs. E A healthy control fetus of the same gestational age. The sagittal-view image of the profile shows a normal maxilla and mandible. F The fetus in our case. The sagittal-view image of the profile shows thick and everted upper and lower lips
After obtaining approval from the Ethics Committee of the First People’s Hospital of Yunnan Province (KHLL2021-169) and receiving written informed consent from each patient, genomic DNA was extracted from the proband, biological parents, and fetal amniotic fluid according to the manufacturer’s instructions. WES was then performed with the MGISEQ-2000 platform (BGI, Shenzhen, China). The workflow comprised DNA sample quality testing, DNA fragment library construction, exon capture, on-machine sequencing, and off-machine data-quality control. The following metrics were achieved for all samples: a mean coverage depth > 200X and interrogation > 95% of target bases (exons ± 30 intronic nucleotides flanking the exon-intron boundaries of all nuclear genes) at > 20× read depth. The raw sequences were then aligned to the human reference genome (hg37) using BWA software. We retrieved disease-causing variants from bioinformatics databases such as the Genome Mutation Frequency Database (gnomAD), Online Mendelian Genetics Database, ClinVar Database, and the Human Gene Mutation Database. PROVEAN (http://provean.jcvi.org/protein_batch_submit.php?species=human), Polyphen2 (http://genetics.bwh.harvard.edu/pph2), and other in silico tools were subsequently adopted to predict whether the variant was considered to be deleterious. The pathogenicity of the variants was interpretated according to the American College of Medical Genetics and Genomics (ACMG) variation classification guidelines, and the candidate variants were validated by Sanger sequencing.
The family pedigree is presented in Fig. 3A. The WES results revealed a hemizygous variant in exon 7 of the EDA gene (NM_001399.4: c.806G > T, p.Gly269Val) in the proband, and the mother was identified as a heterozygous carrier of this variant, while the father did not carry it (Fig. 3B). According to the ACMG guidelines [13], the EDA c.806G > T variant was considered to be likely pathogenic (PM1 + PM2 + PP3 + PP4). Based on the above genetic test results, we used amniotic fluid extracted from the pregnant woman to verify whether the fetus harbored the variant of the EDA gene c.806G > T; and the results showed that the fetus—like the proband—also contained the heterozygous variant at the aforementioned loci (Fig. 3B). This result was consistent with the X-linked recessive inheritance characteristics. Although approximately 411 pathogenic or likely pathogenic HED variants have been listed in the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar), the variant site of the EDA gene in our study was not reported. In addition, the c.806G > T variant in exon 7 of the EDA gene that we identified was not documented in the gnomAD database. The PROVEAN online prediction software classified this novel missense variant as deleterious (score = − 3.347), and this variant was predicted by PolyPhen2 to be probably damaging, with a score of 1. Amino acid sequence alignment revealed that the p.Gly269Val variant was located at highly conserved residues across several mammalian species (Fig. 3C). This high conservation suggests that the site is essential for the normal structure and function of the protein. Based on family history, clinical manifestations, and genetic testing results, we diagnosed the proband and the aborted fetus with HED caused by the novel EDA variant.
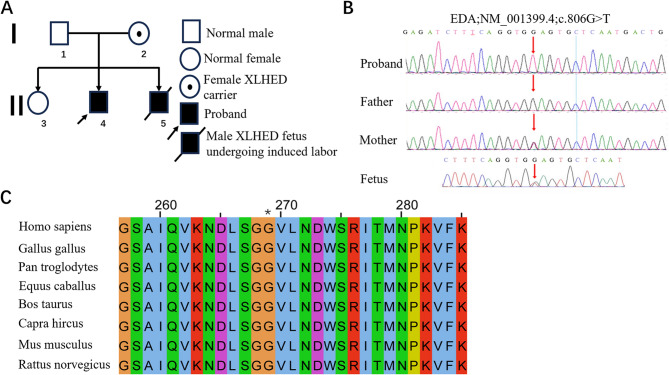
Fig. 3.
A Constructed family tree. Number 5 in line II is the afflicted fetus in this case. B WES identified a hemizygous variation in exon 7 of the EDA gene in the proband (NM_001399.4, c. 806G > T, p.Gly269val), and the mother was a heterozygous carrier for this variation; the father did not carry the variation. Sanger sequencing confirmed that the fetus harbored a heterozygous genotype for the variation. C Evolutionary conservation of amino acid residues as altered by p.Gly269Val across various species. NCBI accession numbers are as follows: Homo sapiens, NP_001005609.1; Gallus gallus, NP_001409628.1; Pan troglodytes, XP_009437490.1; Equus caballus, XP_014584436.2; Bos taurus, NP_001075212.1; Capra hircus, XP_017899470.1; Mus musculus, NP_001171408.1; and Rattus norvegicus, NP_001292172.1. Asterisk (*) denotes 269 Gly
Discussion
We herein analyzed a family using relatively comprehensive clinical data that reflected XLHED. The proband manifested typical clinical characteristics, a prenatal ultrasound examination of the induced male fetus exhibited a characteristic image, and genetic testing showed a variant site that had never been reported before. The family history of the patient also revealed a typical X-linked recessive genetic characteristic.
XLHED is a rare genetic disorder with a prevalence of 2.99 cases per 100,000 live births [14]. Affected individuals manifest abnormal development of their sweat glands, tooth germ tissue, hair follicles, and nails; and they exhibit prominent supraorbital ridges, saddle noses, forehead bulges, hypoplastic nasal wings, and missing teeth [15]. However, there is as yet no effective prenatal diagnostic method for this disease. Although some studies have suggested that ultrasonographic examination constitutes a reliable prenatal diagnostic method for XLHED, only a few authors have reported prenatal ultrasound characteristics of HED fetuses such as the absence of tooth germs or fewer tooth germs [16–18]; a small nose and thick, protruding lips [19]; short and retracted maxillary and mandibular bones [11]; and malocclusion—all of which can readily lead to the conspicuous facial features characteristic of the disorder [20]. While we reported prenatal ultrasound images with similar characteristics in our case of XLHED, due to the limitations of ultrasonographic examination, we experienced insurmountable difficulties in observing the development of sweat glands, hair follicles, and nails in the fetus. In addition, factors such as the physicians’ understanding of the disease, fetal position, and gestational age also affect the detection rate of the disease.
Genetic examination is an important means used to diagnose HED, which is primarily caused by mutations in ectodysplasin A (> 50% cases), EDA receptor (EDAR), EDARADD, and/or WNT10A [12]. Ectodysplasin-A (encoded by the EDA gene) is a member of the tumor necrosis factor (TNF) superfamily of ligands, and it acts as a homotrimer that may be involved in cell-cell signaling during the development of ectodermal organs, a function that is extremely important for producing various structures of ectoderm [21]. We identified a novel c.806G > T variant on exon 7 of the EDA gene that was absent in the ClinVar and gnomAD databases. The pathogenicity of this variant was confirmed by family history, clinical manifestations, database analysis of human genome variation, and functional software prediction. In our review of exon 7 variants (Table 1), we ascertained that all ClinVar-classified pathogenic/likely pathogenic variants caused severe oligodontia, dry skin, and hypohidrosis in males; while female carriers only demonstrated dental anomalies—and this was consistent with our proband. Intriguingly, affected individuals exhibited normal cognition. We posit that this genotype-phenotype correlation will enhance prenatal counseling. However, incomplete knowledge of HED-associated genes limits genetic testing sensitivity and thus poses greater risks of missed diagnoses.
Table 1.
Clinical and Genetic Summary of exon 7 of the EDA gene in XLHED patients
| Variants | Sex | anodontia or oligoodontia | Absent/ sparse hair | Dry skin | Hypohidrosis /Anhidrosis | Intellectual disability | Craniofacial abnormalities | Increased susceptibility to infections | Nose bleed | Molecular consequence | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|
| c.806G>T (p.Gly269val) | M | + | + | + | + | - | + | - | - | missense | Our cases |
| c.794-1G>A | M | + | + | - | + | - | - | - | - | splice acceptor | doi: 10.1086/301984 |
| c.801A>G (p.Ser267=) | M | + | + | + | - | - | + | + | - | synonymous | doi: 10.1111/jdv.15798 |
| c.822del (p.Trp274fs) | M | - | - | + | + | - | + | - | - | frameshift |
doi: 10.1002/mgg3.84 doi: 10.1002/ajmg.a.35959 |
| c.822G>A (p.Trp274Ter) | M | + | + | - | + | - | - | - | - | nonsense | doi: 10.1074/jbc.M101280200 |
| c.826C>T (p.Arg276Cys) | M | + | + | + | + | - | - | - | - | missense | doi: 10.1002/ajmg.a.32994 |
| c.836T>G (p.Met279Arg) | M | + | + | - | + | - | - | - | + | missense | doi:10.1111/j.1365-2133.2007.08231.x |
| c.865C>T (p.Arg289Cys) | M | + | - | - | - | - | - | - | - | missense | doi:10.1007/s00056-015-0005-1 |
| c.866G>A (p.Arg289His) | M | + | + | + | - | - | - | - | - | missense | doi: 10.1002/mgg3.1555 |
| F | + | - | - | - | - | - | - | - | missense | ||
| c.871G>C (p.Gly291Arg) | M | + | + | - | + | - | + | - | - | missense |
doi:10.1002/humu.21384 doi: 10.1007/s00431-013-1985-8 |
| c.871G>A (p.Gly291Arg) | M | + | + | + | + | - | + | - | - | missense |
doi: 10.1002/humu.21384 doi: 10.1159/000500214 |
| c.878T>G (p.Leu293Arg) | M | + | - | - | - | - | - | - | - | missense | doi:10.1186/s12881-018-0726-2 |
| c.896G>A (p.Gly299Asp) | M | + | + | + | + | - | - | - | - | missense | doi:10.1002/humu.21384doi:http://dx doi.org/10.4238/2015.August.28.21 |
| c.902A>G (p.Tyr301Cys) | M | + | + | + | + | - | + | - | - | missense | doi:10.1016/j.ejmg.2011.03.005 |
| c.917A>G (p.Gln306Arg) | M | + | + | + | + | - | + | - | - | missense | doi:10.1111/1346-8138.13479 |
M male, F female
+the patient possessed this clinical manifestation; -not provided
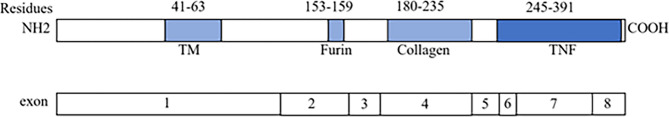
Ectodysplasin-A has a transmembrane domain, a putative furin cleavage site, a collagen subdomain, and a TNF homology domain (THD) [22]. The THD mediates receptor binding through cysteine-rich domain (CRD) interactions with EDAR/XEDAR, activating NF-κB signaling that is essential for ectodermal morphogenesis [4]. Notably, a large number of variants were located in the TNF homology domain [23], a pattern that was corroborated by our findings, i.e., the variant of the EDA gene c.806G > T localized to the exon 7-encoded THD (Fig. 4). This substitution likely disrupts ligand-receptor binding or downstream signal transduction, offering a mechanistic explanation for the observed XLHED phenotype.
Fig. 4.
Schematic drawing of the EDA gene showing exonic structure and predicted protein motifs. TM, transmembrane domain; furin, furin cleavage site; collagen, collagen-like domain; TNF, tumor necrosis factor homology domain. We found the predicted protein motif information in Park, J Setal. 1996; and exonic data are from the UCSC Genome Browser (https://genome.ucsc.edu)
While XLHED is associated with a 30% mortality rate in childhood that is principally attributable to recurrent infectious complications [24], current therapies for EDA deficiency remain unproven. Fc-EDA, a recombinant fusion protein that encompasses ectodysplasin-A’s receptor-binding domain and IgG1-Fc, shows therapeutic promise; and in murine models, prenatal Fc-EDA administration prevented XLHED manifestations. Human trials have also demonstrated safety. For example, intra-amniotic Fc-EDA administered at 26–31 gestational weeks to male fetuses yielded normal gland development by 14–22 months, with no anti-drug antibodies detected in either mothers or infants [16, 25]. These findings highlight Fc-EDA’s potential and reinforce the importance of detecting XLHED in prenatal diagnosis.
Conclusions
The prenatal ultrasound evaluation of tooth germ development offers valuable evidence for prenatal genetic testing and potentially influences the outcome of pregnancy. Notably, this case involved a family history of HED, prenatal diagnosis is essential. In this study, we identified a previously unreported de novo c.806G > T variant in the EDA gene in a fetus from such a high-risk family. The findings of this study expand the existing spectrum of EDA gene mutations and offer assistance in genetic counseling, prenatal diagnosis, and intervention for families affected by XLHED.
Supplementary Information
Acknowledgements
Thanks to LetPub (www.letpub.com.cn) for its linguistic assistance during the preparation of this manuscript, to the pregnant couple for their support, and to each of the authors.
Abbreviations
- EDA gene
Ectodysplasin A gene
- HED
Hypohidrotic ectodermal dysplasia
- WES
Whole exome sequencing
- NT
Nuchal Translucency
Author contributions
LM Y diagnosed the case and sorted out the data, LM Y and LL W conducted article writing, JM Z and BS Z diagnosed the case and review the case report, YH L collected the proband’s history, YH Z managed the patient and obtained the patient’s history, XL C and LM Y con-ducted anatomical verification, and JH Y followed up the case, GY H provided language service. All authors have read and approved the manuscript.
Funding
1. National Natural Area Foundation Project: Study on the association and role of key intestinal flora and its metabolites with the risk of preeclampsia based on birth cohort. Project Approval Number: 82260645.
2.Doctor Foundation of the First People’s Hospital of Yunnan province, Grant/Award Number: KHBS-2022-016;
3.Yunnan Provincial Department of Science and Technology - Kunming Medical University Joint Special Project on Applied Basic Research, Grant/Award Number: (202201AY070001-226)
Data availability
Data is provided within the manuscript or supplementary information files.
Declarations
Ethics approval and consent to participate
Written informed consent was obtained from the patient and her husband. The study was approved by the Ethics Committee of the First People’s Hospital of Yunnan Province (KHLL2021-169).
Consent for publication
We have obtained the consent of all participants with personal information involved in the case and signed the consent for publication. A copy of the signed, written informed consent for publication form is available for review by the editor.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Limin Yao and Lilan Wan contributed equally to this work.
References
- 1.Agarwal S, Gupta S. Hypohidrotic ectodermal dysplasia. INDIAN DERMATOL ONL. 2012;3(2):125–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bansal M, Manchanda K, Pandey SS. Hypohidrotic ectodermal dysplasia. Int J Trichology. 2012;4(3):167–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cui CY, Schlessinger D. EDA signaling and skin appendage development. Cell Cycle. 2006;5(21):2477–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu K, Huang C, Wan F, Jiang C, Chen J, Li X, et al. Structural insights into pathogenic mechanism of hypohidrotic ectodermal dysplasia caused by Ectodysplasin A variants. Nat Commun. 2023;14(1):767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monreal AW, Ferguson BM, Headon DJ, Street SL, Overbeek PA, Zonana J. Mutations in the human homologue of mouse Dl cause autosomal recessive and dominant hypohidrotic ectodermal dysplasia. Nat Genet. 1999;22(4):366–9. [DOI] [PubMed] [Google Scholar]
- 6.Duverger O, Lee D, Hassan MQ, Chen SX, Jaisser F, Lian JB, et al. Molecular consequences of a frameshifted DLX3 mutant leading to Tricho-Dento-Osseous syndrome. J Biol Chem. 2008;283(29):20198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nieminen P, Lukinmaa PL, Alapulli H, Methuen M, Suojärvi T, Kivirikko S, et al. DLX3 homeodomain mutations cause tricho-dento-osseous syndrome with novel phenotypes. Cells Tissues Organs. 2011;194(1):49–59. [DOI] [PubMed] [Google Scholar]
- 8.Adaimy L, Chouery E, Megarbane H, Mroueh S, Delague V, Nicolas E, et al. Mutation in WNT10A is associated with an autosomal recessive ectodermal dysplasia: the odonto-onycho-dermal dysplasia. AM J HUM GENET. 2007;81(4):821–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pigno MA, Blackman RB, Cronin RJ, Cavazos E. Prosthodontic management of ectodermal dysplasia: a review of the literature. J PROSTHET DENT. 1996;76(5):541–5. [DOI] [PubMed] [Google Scholar]
- 10.Hammersen J, Wohlfart S, Goecke TW, Köninger A, Stepan H, Gallinat R, et al. Reliability of prenatal detection of X-linked hypohidrotic ectodermal dysplasia by tooth germ sonography. PRENATAL DIAG. 2018;39(9):796–805. [DOI] [PubMed] [Google Scholar]
- 11.Li L, Zhou Y, Tian R, Zhang C. Prenatal ultrasound findings of ectodermal dysplasia: a case report. BMC Pregnancy Childbirth. 2022;22(1):100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bagdey SP, Moharil RB, Dive A, Bodhade A. Hypohidrotic ectodermal dysplasia: A case report with review and latest updates. J Oral Maxillofac Pathol. 2022;26(Suppl 1):S12-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. GENET MED. 2015;17(5):405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butcher C, Abbott BM, Grange D, Fete M, Meyer B, Spinka C, et al. Prevalence rates for ectodermal dysplasia syndromes. AM J MED GENET A. 2024;194(12):e63832. [DOI] [PubMed] [Google Scholar]
- 15.de Aquino SN, Paranaíba LM, Swerts MS, Martelli DR, de Barros LM. Martelli júnior H. Orofacial features of hypohidrotic ectodermal dysplasia. HEAD NECK PATHOL. 2012;6(4):460–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider H, Faschingbauer F, Schuepbach-Mallepell S, Körber I, Wohlfart S, Dick A, et al. Prenatal correction of X-Linked hypohidrotic ectodermal dysplasia. NEW ENGL J MED. 2018;378(17):1604–10. [DOI] [PubMed] [Google Scholar]
- 17.Duan F, Wang C, Ren S, Kong X. Prenatal diagnosis of a fetus with X-linked hypohidrotic ectodermal dysplasia. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2020;37(11):1269–71. [DOI] [PubMed] [Google Scholar]
- 18.Wünsche S, Jüngert J, Faschingbauer F, Mommsen H, Goecke T, Schwanitz K, et al. Noninvasive prenatal diagnosis of hypohidrotic ectodermal dysplasia by tooth germ sonography. ULTRASCHALL MED. 2014;36(4):381–5. [DOI] [PubMed] [Google Scholar]
- 19.Sepulveda W, Sandoval R, Carstens E, Gutierrez J, Vasquez P. Hypohidrotic ectodermal dysplasia: prenatal diagnosis by three-dimensional ultrasonography. J ULTRAS MED. 2003;22(7):731–5. [DOI] [PubMed] [Google Scholar]
- 20.Li TG, Ma B, Tie HX, Zhang QH, Hao SJ, Guan CL. Prenatal sonographic diagnosis of X-linked hypohidrotic ectodermal dysplasia: an unusual case. J CLIN ULTRASOUND. 2021;49(8):838–40. [DOI] [PubMed] [Google Scholar]
- 21.Anbouba GM, Carmany EP, Natoli JL. The characterization of hypodontia, hypohidrosis, and hypotrichosis associated with X-linked hypohidrotic ectodermal dysplasia: A systematic review. AM J MED GENET A. 2020;182(4):831–41. [DOI] [PubMed] [Google Scholar]
- 22.Huang SX, Liang JL, Sui WG, Lin H, Xue W, Chen JJ, et al. EDA mutation as a cause of hypohidrotic ectodermal dysplasia: a case report and review of the literature. Genet Mol Res. 2015;14(3):10344–51. [DOI] [PubMed] [Google Scholar]
- 23.Han Y, Wang X, Zheng L, Zhu T, Li Y, Hong J, et al. Pathogenic EDA mutations in Chinese Han families with hypohidrotic ectodermal dysplasia and genotype-phenotype: a correlation analysis. Front Genet. 2020;11: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fete M, Hermann J, Behrens J, Huttner KM. X-linked hypohidrotic ectodermal dysplasia (xlhed): clinical and diagnostic insights from an international patient registry. Am J Med Genet A. 2014;164A(10):2437–42. [DOI] [PubMed] [Google Scholar]
- 25.Körber I, Klein OD, Morhart P, Faschingbauer F, Grange DK, Clarke A, et al. Safety and immunogenicity of Fc-EDA, a Recombinant Ectodysplasin A1 replacement protein, in human subjects. BRIT J CLIN PHARMACO. 2020;86(10):2063–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is provided within the manuscript or supplementary information files.