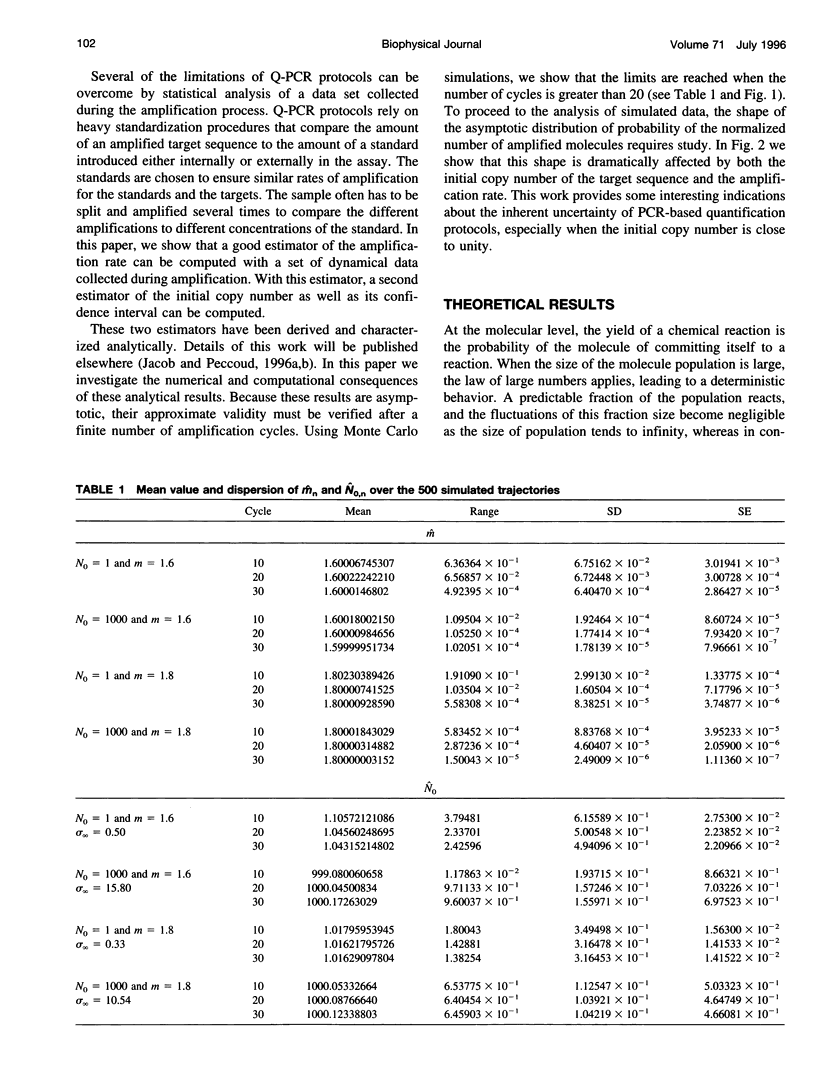
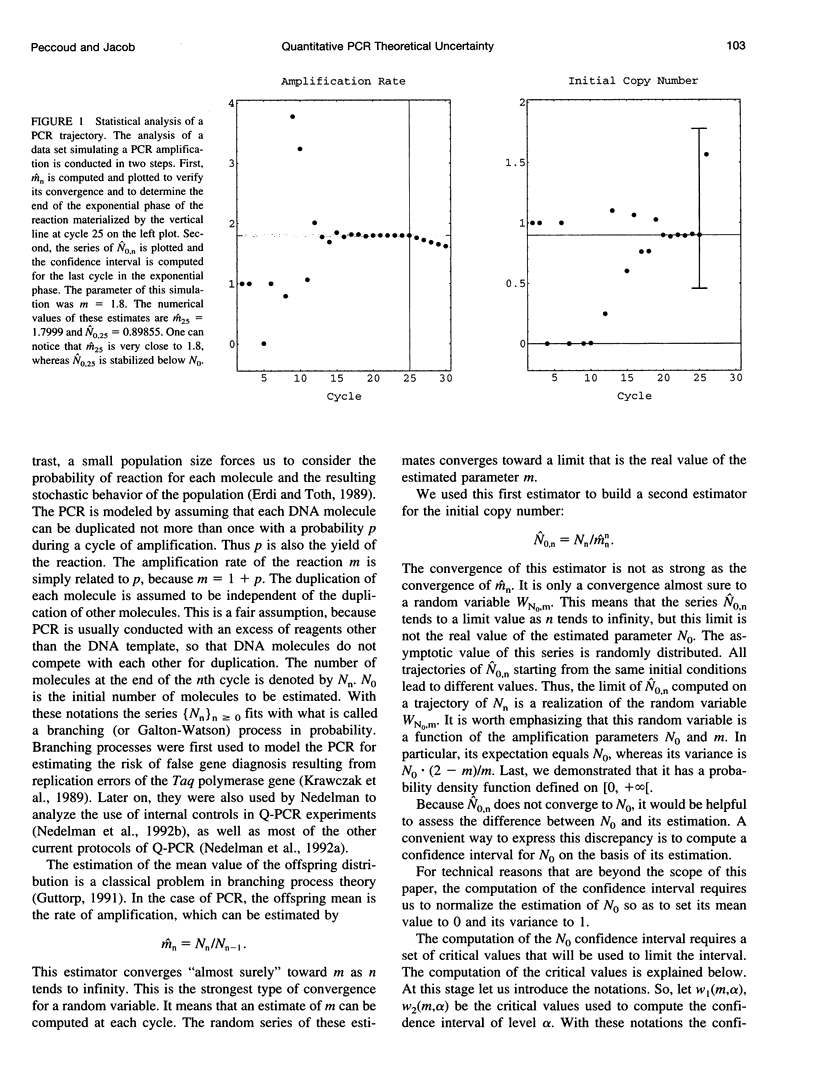
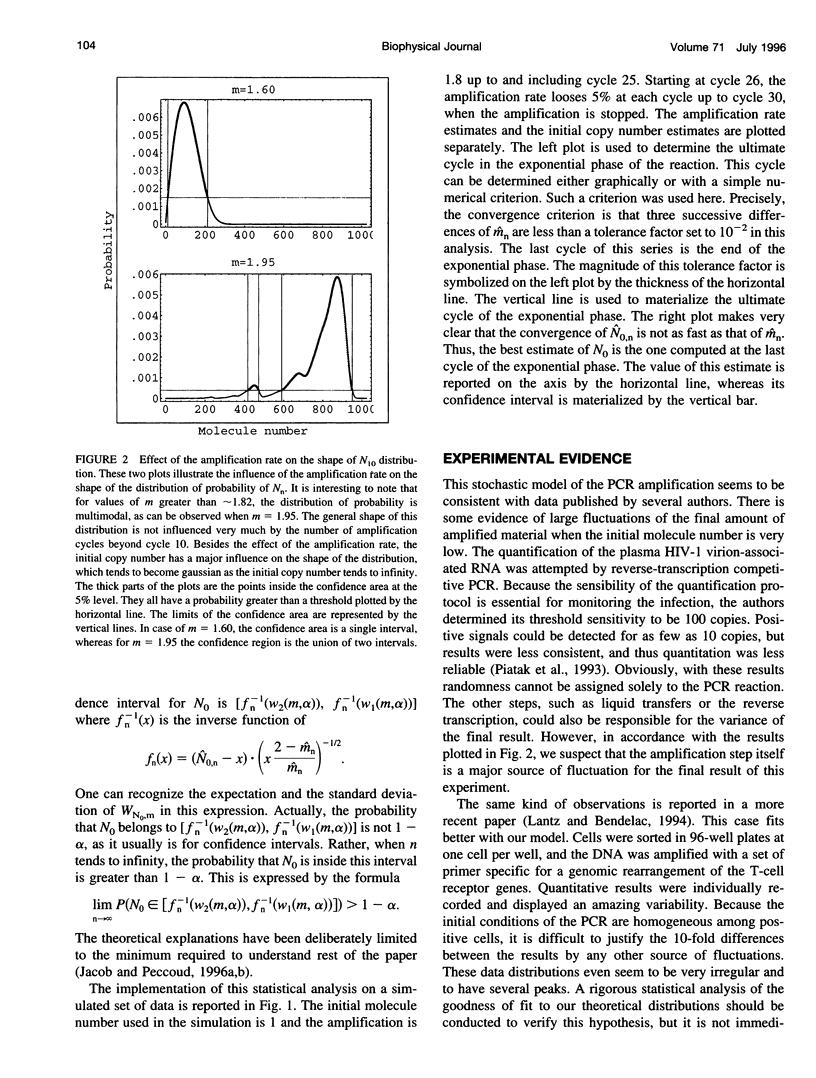
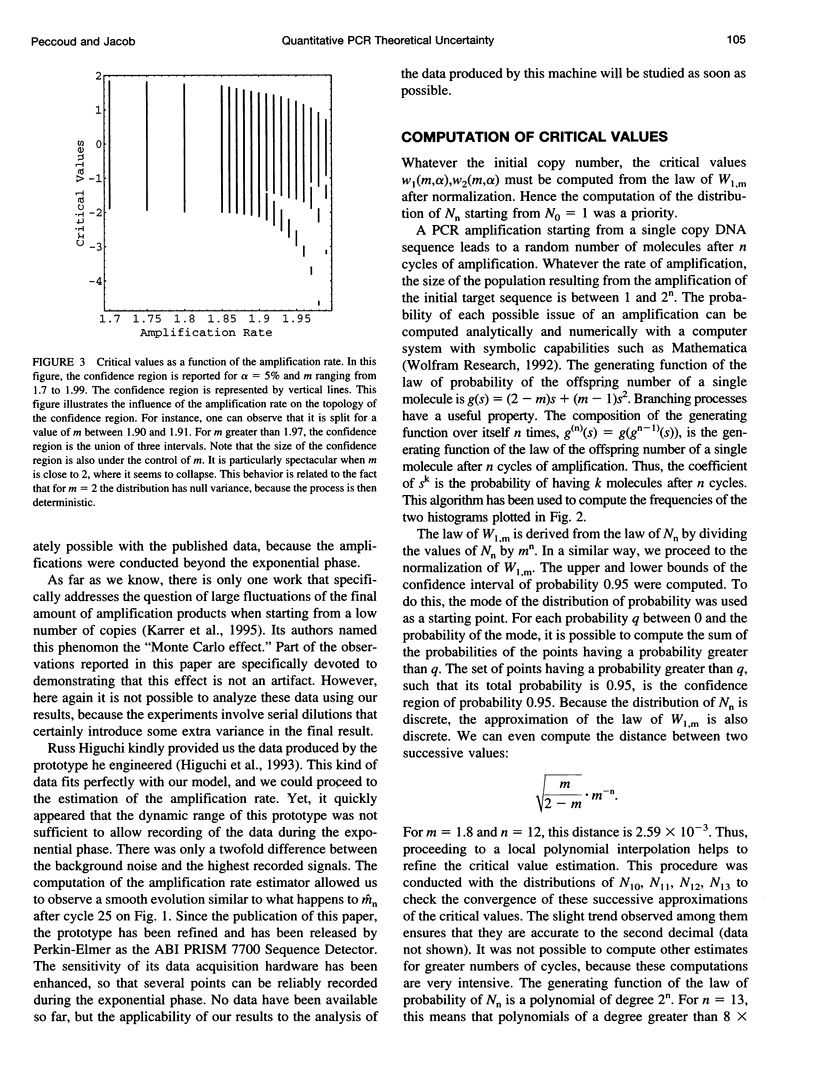
Abstract
Current quantitative polymerase chain reaction (PCR) protocols are only indicative of the quantity of a target sequence relative to a standard, because no means of estimating the amplification rate is yet available. The variability of PCR performed on isolated cells has already been reported by several authors, but it could not be extensively studied, because of lack of a system for doing kinetic data acquisition and of statistical methods suitable for analyzing this type of data. We used the branching process theory to simulate and analyze quantitative kinetic PCR data. We computed the probability distribution of the offspring of a single molecule. We demonstrated that the rate of amplication has a severe influence on the shape of this distribution. For high values of the amplification rate, the distribution has several maxima of probability. A single amplification trajectory is used to estimate the initial copy number of the target sequence as well as its confidence interval, provided that the amplification is done over more than 20 cycles. The consequence of possible molecular fluctuations in the early stage of amplification is that small copy numbers result in relatively larger intervals than large initial copy numbers. The confidence interval amplitude is the theoretical uncertainty of measurements using quantitative PCR. We expect these results to be applicable to the data produced by the next generation of thermocyclers for quantitative applications.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cui X. F., Li H. H., Goradia T. M., Lange K., Kazazian H. H., Jr, Galas D., Arnheim N. Single-sperm typing: determination of genetic distance between the G gamma-globin and parathyroid hormone loci by using the polymerase chain reaction and allele-specific oligomers. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9389–9393. doi: 10.1073/pnas.86.23.9389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlich H. A., Gelfand D., Sninsky J. J. Recent advances in the polymerase chain reaction. Science. 1991 Jun 21;252(5013):1643–1651. doi: 10.1126/science.2047872. [DOI] [PubMed] [Google Scholar]
- Higuchi R., Fockler C., Dollinger G., Watson R. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology (N Y) 1993 Sep;11(9):1026–1030. doi: 10.1038/nbt0993-1026. [DOI] [PubMed] [Google Scholar]
- Hoof T., Riordan J. R., Tümmler B. Quantitation of mRNA by the kinetic polymerase chain reaction assay: a tool for monitoring P-glycoprotein gene expression. Anal Biochem. 1991 Jul;196(1):161–169. doi: 10.1016/0003-2697(91)90133-e. [DOI] [PubMed] [Google Scholar]
- Karrer E. E., Lincoln J. E., Hogenhout S., Bennett A. B., Bostock R. M., Martineau B., Lucas W. J., Gilchrist D. G., Alexander D. In situ isolation of mRNA from individual plant cells: creation of cell-specific cDNA libraries. Proc Natl Acad Sci U S A. 1995 Apr 25;92(9):3814–3818. doi: 10.1073/pnas.92.9.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczak M., Reiss J., Schmidtke J., Rösler U. Polymerase chain reaction: replication errors and reliability of gene diagnosis. Nucleic Acids Res. 1989 Mar 25;17(6):2197–2201. doi: 10.1093/nar/17.6.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantz O., Bendelac A. An invariant T cell receptor alpha chain is used by a unique subset of major histocompatibility complex class I-specific CD4+ and CD4-8- T cells in mice and humans. J Exp Med. 1994 Sep 1;180(3):1097–1106. doi: 10.1084/jem.180.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. H., Gyllensten U. B., Cui X. F., Saiki R. K., Erlich H. A., Arnheim N. Amplification and analysis of DNA sequences in single human sperm and diploid cells. Nature. 1988 Sep 29;335(6189):414–417. doi: 10.1038/335414a0. [DOI] [PubMed] [Google Scholar]
- Lion T., Henn T., Gaiger A., Kalhs P., Gadner H. Early detection of relapse after bone marrow transplantation in patients with chronic myelogenous leukaemia. Lancet. 1993 Jan 30;341(8840):275–276. doi: 10.1016/0140-6736(93)92619-5. [DOI] [PubMed] [Google Scholar]
- Nedelman J., Heagerty P., Lawrence C. Quantitative PCR with internal controls. Comput Appl Biosci. 1992 Feb;8(1):65–70. doi: 10.1093/bioinformatics/8.1.65. [DOI] [PubMed] [Google Scholar]
- Piatak M., Jr, Saag M. S., Yang L. C., Clark S. J., Kappes J. C., Luk K. C., Hahn B. H., Shaw G. M., Lifson J. D. High levels of HIV-1 in plasma during all stages of infection determined by competitive PCR. Science. 1993 Mar 19;259(5102):1749–1754. doi: 10.1126/science.8096089. [DOI] [PubMed] [Google Scholar]
- Raeymaekers L. Comments on quantitative PCR. Eur Cytokine Netw. 1994 Jan-Feb;5(1):57–60. [PubMed] [Google Scholar]
- Raeymaekers L. Quantitative PCR: theoretical considerations with practical implications. Anal Biochem. 1993 Nov 1;214(2):582–585. doi: 10.1006/abio.1993.1542. [DOI] [PubMed] [Google Scholar]
- Santagati S., Bettini E., Asdente M., Muramatsu M., Maggi A. Theoretical considerations for the application of competitive polymerase chain reaction to the quantitation of a low abundance mRNA: estrogen receptor. Biochem Pharmacol. 1993 Nov 17;46(10):1797–1803. doi: 10.1016/0006-2952(93)90585-k. [DOI] [PubMed] [Google Scholar]
- Semple M. G., Kaye S., Loveday C., Tedder R. S. HIV-1 plasma viraemia quantification: a non-culture measurement needed for therapeutic trials. J Virol Methods. 1993 Feb;41(2):167–179. doi: 10.1016/0166-0934(93)90124-a. [DOI] [PubMed] [Google Scholar]
- Simmonds P., Zhang L. Q., Watson H. G., Rebus S., Ferguson E. D., Balfe P., Leadbetter G. H., Yap P. L., Peutherer J. F., Ludlam C. A. Hepatitis C quantification and sequencing in blood products, haemophiliacs, and drug users. Lancet. 1990 Dec 15;336(8729):1469–1472. doi: 10.1016/0140-6736(90)93179-s. [DOI] [PubMed] [Google Scholar]
- Singer-Sam J., Chapman V., LeBon J. M., Riggs A. D. Parental imprinting studied by allele-specific primer extension after PCR: paternal X chromosome-linked genes are transcribed prior to preferential paternal X chromosome inactivation. Proc Natl Acad Sci U S A. 1992 Nov 1;89(21):10469–10473. doi: 10.1073/pnas.89.21.10469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snabes M. C., Chong S. S., Subramanian S. B., Kristjansson K., DiSepio D., Hughes M. R. Preimplantation single-cell analysis of multiple genetic loci by whole-genome amplification. Proc Natl Acad Sci U S A. 1994 Jun 21;91(13):6181–6185. doi: 10.1073/pnas.91.13.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. M., Doyle M. V., Mark D. F. Quantitation of mRNA by the polymerase chain reaction. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9717–9721. doi: 10.1073/pnas.86.24.9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner R. J., Beinbrech B., Rüegg J. C. Quantitative PCR. Nature. 1993 Dec 2;366(6454):416–416. doi: 10.1038/366416b0. [DOI] [PubMed] [Google Scholar]
- Wiesner R. J., Rüegg J. C., Morano I. Counting target molecules by exponential polymerase chain reaction: copy number of mitochondrial DNA in rat tissues. Biochem Biophys Res Commun. 1992 Mar 16;183(2):553–559. doi: 10.1016/0006-291x(92)90517-o. [DOI] [PubMed] [Google Scholar]
- Wood R., Dong H., Katzenstein D. A., Merigan T. C. Quantification and comparison of HIV-1 proviral load in peripheral blood mononuclear cells and isolated CD4+ T cells. J Acquir Immune Defic Syndr. 1993 Mar;6(3):237–240. [PubMed] [Google Scholar]
- Zhang L., Cui X., Schmitt K., Hubert R., Navidi W., Arnheim N. Whole genome amplification from a single cell: implications for genetic analysis. Proc Natl Acad Sci U S A. 1992 Jul 1;89(13):5847–5851. doi: 10.1073/pnas.89.13.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]



