Abstract
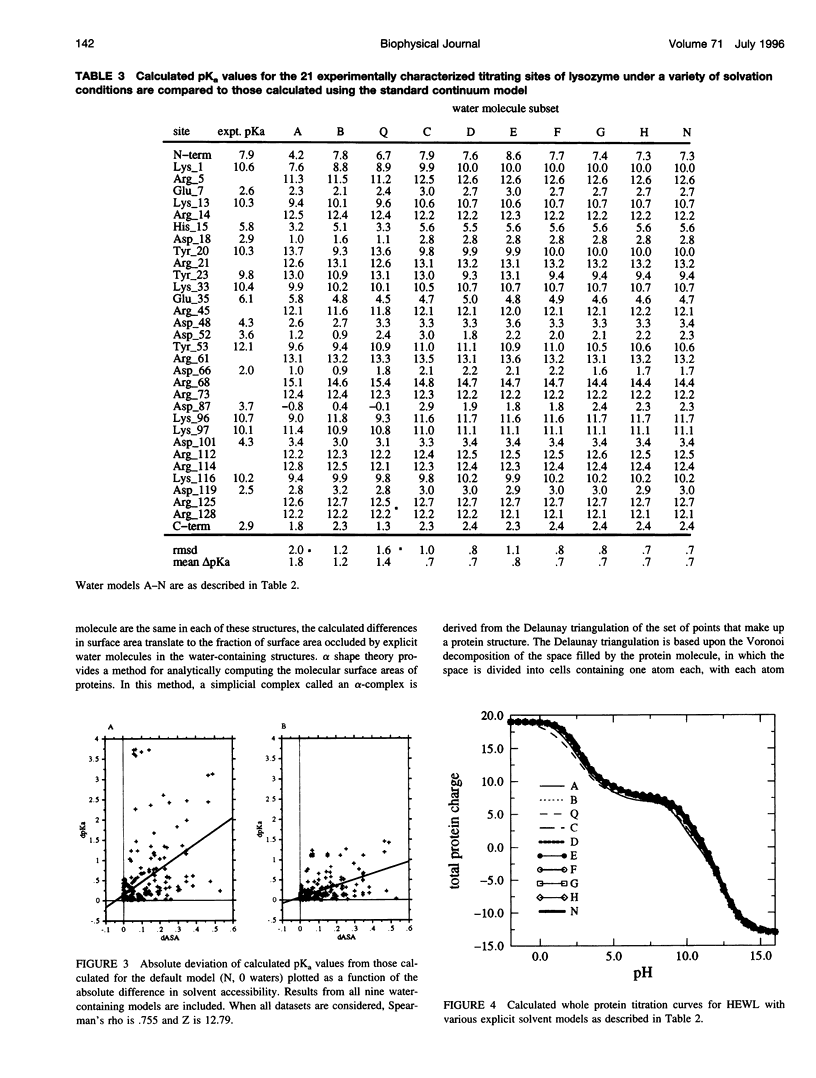
Continuum methods for calculation of protein electrostatics treat buried and ordered water molecules by one of two approximations; either the dielectric constant of regions containing ordered water molecules is equal to the bulk solvent dielectric constant, or it is equal to the protein dielectric constant though no fixed atoms are used to represent water molecules. A method for calculating the titration behavior of individual residues in proteins has been tested on models of hen egg white lysozyme containing various numbers of explicit water molecules. Water molecules were included based on hydrogen bonding, solvent accessibility, and/or proximity to titrating groups in the protein. Inclusion of water molecules significantly alters the calculated titration behavior of individual titrating sites, shifting calculated pKa values by up to 0.5 pH unit. Our results suggest that approximately one water molecule within hydrogen-bonding distance of each charged group should be included in protein electrostatics calculations.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antosiewicz J., McCammon J. A., Gilson M. K. Prediction of pH-dependent properties of proteins. J Mol Biol. 1994 May 6;238(3):415–436. doi: 10.1006/jmbi.1994.1301. [DOI] [PubMed] [Google Scholar]
- Bashford D., Karplus M. pKa's of ionizable groups in proteins: atomic detail from a continuum electrostatic model. Biochemistry. 1990 Nov 6;29(44):10219–10225. doi: 10.1021/bi00496a010. [DOI] [PubMed] [Google Scholar]
- Beroza P., Fredkin D. R., Okamura M. Y., Feher G. Protonation of interacting residues in a protein by a Monte Carlo method: application to lysozyme and the photosynthetic reaction center of Rhodobacter sphaeroides. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5804–5808. doi: 10.1073/pnas.88.13.5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A. T., Karplus M. Polar hydrogen positions in proteins: empirical energy placement and neutron diffraction comparison. Proteins. 1988;4(2):148–156. doi: 10.1002/prot.340040208. [DOI] [PubMed] [Google Scholar]
- Colombo M. F., Rau D. C., Parsegian V. A. Protein solvation in allosteric regulation: a water effect on hemoglobin. Science. 1992 May 1;256(5057):655–659. doi: 10.1126/science.1585178. [DOI] [PubMed] [Google Scholar]
- Gilson M. K. Multiple-site titration and molecular modeling: two rapid methods for computing energies and forces for ionizable groups in proteins. Proteins. 1993 Mar;15(3):266–282. doi: 10.1002/prot.340150305. [DOI] [PubMed] [Google Scholar]
- Karshikoff A. A simple algorithm for the calculation of multiple site titration curves. Protein Eng. 1995 Mar;8(3):243–248. doi: 10.1093/protein/8.3.243. [DOI] [PubMed] [Google Scholar]
- Kuramitsu S., Hamaguchi K. Analysis of the acid-base titration curve of hen lysozyme. J Biochem. 1980 Apr;87(4):1215–1219. [PubMed] [Google Scholar]
- Lesser G. J., Rose G. D. Hydrophobicity of amino acid subgroups in proteins. Proteins. 1990;8(1):6–13. doi: 10.1002/prot.340080104. [DOI] [PubMed] [Google Scholar]
- McDonald I. K., Thornton J. M. Satisfying hydrogen bonding potential in proteins. J Mol Biol. 1994 May 20;238(5):777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- Meyer E. Internal water molecules and H-bonding in biological macromolecules: a review of structural features with functional implications. Protein Sci. 1992 Dec;1(12):1543–1562. doi: 10.1002/pro.5560011203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberoi H., Allewell N. M. Multigrid solution of the nonlinear Poisson-Boltzmann equation and calculation of titration curves. Biophys J. 1993 Jul;65(1):48–55. doi: 10.1016/S0006-3495(93)81032-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe S. M., Teeter M. M. Patterns for prediction of hydration around polar residues in proteins. J Mol Biol. 1993 Jan 20;229(2):419–427. doi: 10.1006/jmbi.1993.1043. [DOI] [PubMed] [Google Scholar]
- Rousso I., Brodsky I., Lewis A., Sheves M. The role of water in retinal complexation to bacterio-opsin. J Biol Chem. 1995 Jun 9;270(23):13860–13868. doi: 10.1074/jbc.270.23.13860. [DOI] [PubMed] [Google Scholar]
- Tanford C., Roxby R. Interpretation of protein titration curves. Application to lysozyme. Biochemistry. 1972 May 23;11(11):2192–2198. doi: 10.1021/bi00761a029. [DOI] [PubMed] [Google Scholar]
- Wallace A. C., Laskowski R. A., Thornton J. M. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995 Feb;8(2):127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- Williams M. A., Goodfellow J. M., Thornton J. M. Buried waters and internal cavities in monomeric proteins. Protein Sci. 1994 Aug;3(8):1224–1235. doi: 10.1002/pro.5560030808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A. S., Gunner M. R., Sampogna R., Sharp K., Honig B. On the calculation of pKas in proteins. Proteins. 1993 Mar;15(3):252–265. doi: 10.1002/prot.340150304. [DOI] [PubMed] [Google Scholar]







