Abstract
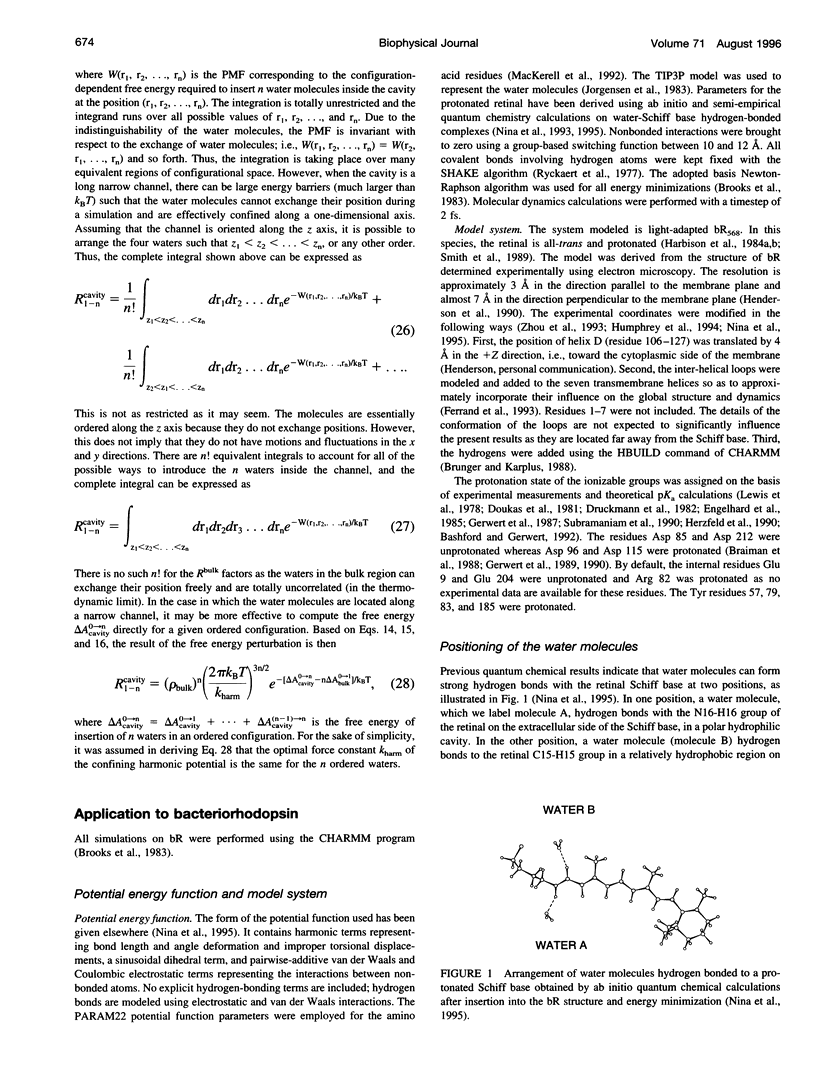
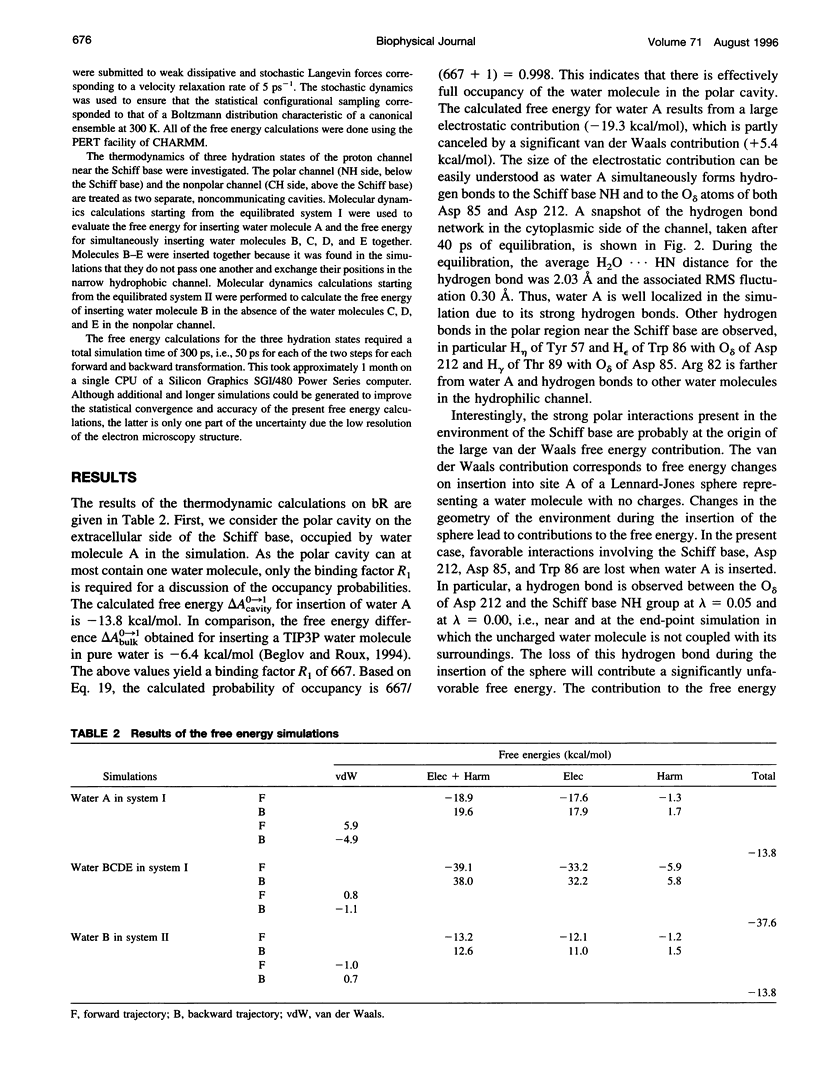
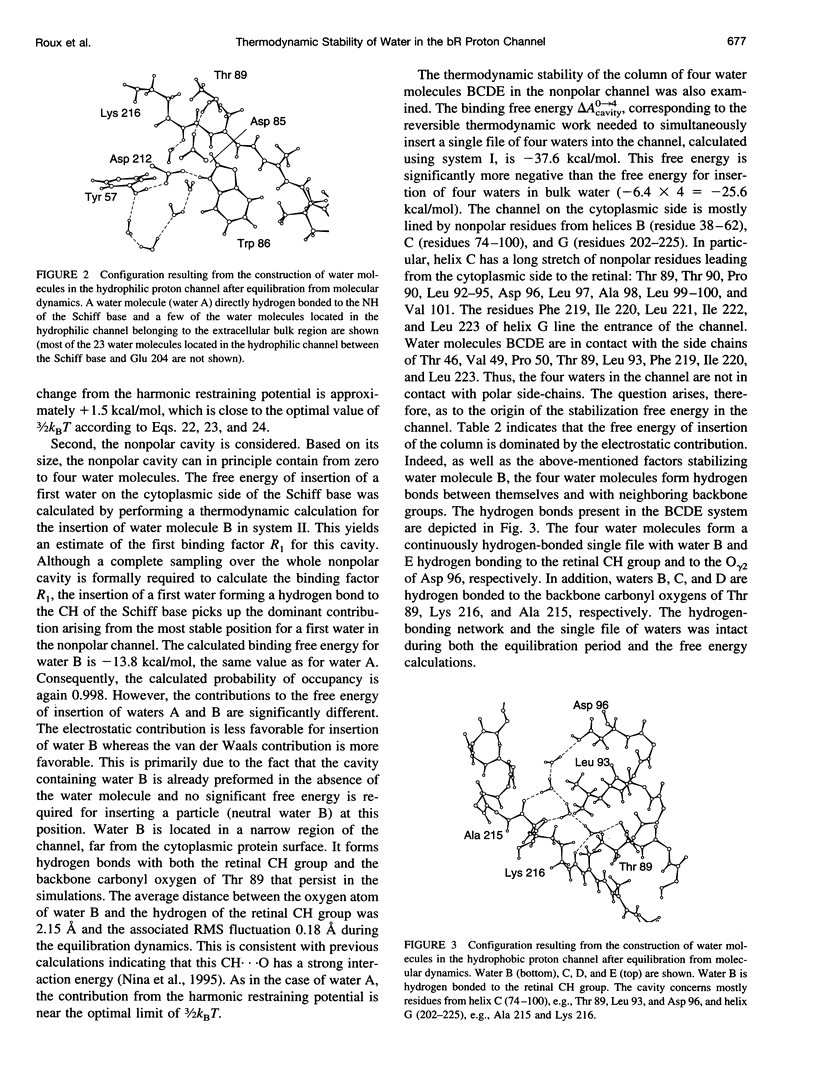
The proton transfer activity of the light-driven proton pump, bacteriorhodopsin (bR) in the photochemical cycle might imply internal water molecules. The free energy of inserting water molecules in specific sites along the bR transmembrane channel has been calculated using molecular dynamics simulations based on a microscopic model. The existence of internal hydration is related to the free energy change on transfer of a water molecule from bulk solvent into a specific binding site. Thermodynamic integration and perturbation methods were used to calculate free energies of hydration for each hydrated model from molecular dynamics simulations of the creation of water molecules into specific protein-binding sites. A rigorous statistical mechanical formulation allowing the calculation of the free energy of transfer of water molecules from the bulk to a protein cavity is used to estimate the probabilities of occupancy in the putative bR proton channel. The channel contains a region lined primarily by nonpolar side-chains. Nevertheless, the results indicate that the transfer of four water molecules from bulk water to this apparently hydrophobic region is thermodynamically permitted. The column forms a continuous hydrogen-bonded chain over 12 A between a proton donor, Asp 96, and the retinal Schiff base acceptor. The presence of two water molecules in direct hydrogen-bonding association with the Schiff base is found to be strongly favorable thermodynamically. The implications of these results for the mechanism of proton transfer in bR are discussed.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bashford D., Gerwert K. Electrostatic calculations of the pKa values of ionizable groups in bacteriorhodopsin. J Mol Biol. 1992 Mar 20;224(2):473–486. doi: 10.1016/0022-2836(92)91009-e. [DOI] [PubMed] [Google Scholar]
- Boresch S., Archontis G., Karplus M. Free energy simulations: the meaning of the individual contributions from a component analysis. Proteins. 1994 Sep;20(1):25–33. doi: 10.1002/prot.340200105. [DOI] [PubMed] [Google Scholar]
- Braiman M. S., Mogi T., Marti T., Stern L. J., Khorana H. G., Rothschild K. J. Vibrational spectroscopy of bacteriorhodopsin mutants: light-driven proton transport involves protonation changes of aspartic acid residues 85, 96, and 212. Biochemistry. 1988 Nov 15;27(23):8516–8520. doi: 10.1021/bi00423a002. [DOI] [PubMed] [Google Scholar]
- Brünger A. T., Karplus M. Polar hydrogen positions in proteins: empirical energy placement and neutron diffraction comparison. Proteins. 1988;4(2):148–156. doi: 10.1002/prot.340040208. [DOI] [PubMed] [Google Scholar]
- Butt H. J., Fendler K., Bamberg E., Tittor J., Oesterhelt D. Aspartic acids 96 and 85 play a central role in the function of bacteriorhodopsin as a proton pump. EMBO J. 1989 Jun;8(6):1657–1663. doi: 10.1002/j.1460-2075.1989.tb03556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Váró G., Chang M., Ni B. F., Needleman R., Lanyi J. K. Water is required for proton transfer from aspartate-96 to the bacteriorhodopsin Schiff base. Biochemistry. 1991 Nov 12;30(45):10972–10979. doi: 10.1021/bi00109a023. [DOI] [PubMed] [Google Scholar]
- Deng H., Huang L., Callender R., Ebrey T. Evidence for a bound water molecule next to the retinal Schiff base in bacteriorhodopsin and rhodopsin: a resonance Raman study of the Schiff base hydrogen/deuterium exchange. Biophys J. 1994 Apr;66(4):1129–1136. doi: 10.1016/S0006-3495(94)80893-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisov V. P., Halle B., Peters J., Hörlein H. D. Residence times of the buried water molecules in bovine pancreatic trypsin inhibitor and its G36S mutant. Biochemistry. 1995 Jul 18;34(28):9046–9051. doi: 10.1021/bi00028a013. [DOI] [PubMed] [Google Scholar]
- Doukas A. G., Pande A., Suzuki T., Callender R. H., Honig B., Ottolenghi M. On the mechanism of hydrogen-deuterium exchange in bacteriorhodopsin. Biophys J. 1981 Feb;33(2):275–279. doi: 10.1016/S0006-3495(81)84889-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druckmann S., Ottolenghi M., Pande A., Pande J., Callender R. H. Acid-base equilibrium of the Schiff base in bacteriorhodopsin. Biochemistry. 1982 Sep 28;21(20):4953–4959. doi: 10.1021/bi00263a019. [DOI] [PubMed] [Google Scholar]
- Edholm O., Berger O., Jähnig F. Structure and fluctuations of bacteriorhodopsin in the purple membrane: a molecular dynamics study. J Mol Biol. 1995 Jun 30;250(1):94–111. doi: 10.1006/jmbi.1995.0361. [DOI] [PubMed] [Google Scholar]
- Edsall J. T., McKenzie H. A. Water and proteins. II. The location and dynamics of water in protein systems and its relation to their stability and properties. Adv Biophys. 1983;16:53–183. doi: 10.1016/0065-227x(83)90008-4. [DOI] [PubMed] [Google Scholar]
- Engelhard M., Gerwert K., Hess B., Kreutz W., Siebert F. Light-driven protonation changes of internal aspartic acids of bacteriorhodopsin: an investigation by static and time-resolved infrared difference spectroscopy using [4-13C]aspartic acid labeled purple membrane. Biochemistry. 1985 Jan 15;24(2):400–407. doi: 10.1021/bi00323a024. [DOI] [PubMed] [Google Scholar]
- Ferrand M., Zaccai G., Nina M., Smith J. C., Etchebest C., Roux B. Structure and dynamics of bacteriorhodopsin. Comparison of simulation and experiment. FEBS Lett. 1993 Aug 2;327(3):256–260. doi: 10.1016/0014-5793(93)80999-b. [DOI] [PubMed] [Google Scholar]
- Fischer W. B., Sonar S., Marti T., Khorana H. G., Rothschild K. J. Detection of a water molecule in the active-site of bacteriorhodopsin: hydrogen bonding changes during the primary photoreaction. Biochemistry. 1994 Nov 1;33(43):12757–12762. doi: 10.1021/bi00209a005. [DOI] [PubMed] [Google Scholar]
- Gerwert K., Hess B., Soppa J., Oesterhelt D. Role of aspartate-96 in proton translocation by bacteriorhodopsin. Proc Natl Acad Sci U S A. 1989 Jul;86(13):4943–4947. doi: 10.1073/pnas.86.13.4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerwert K., Souvignier G., Hess B. Simultaneous monitoring of light-induced changes in protein side-group protonation, chromophore isomerization, and backbone motion of bacteriorhodopsin by time-resolved Fourier-transform infrared spectroscopy. Proc Natl Acad Sci U S A. 1990 Dec 15;87(24):9774–9778. doi: 10.1073/pnas.87.24.9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison G. S., Smith S. O., Pardoen J. A., Mulder P. P., Lugtenburg J., Herzfeld J., Mathies R., Griffin R. G. Solid-state 13C NMR studies of retinal in bacteriorhodopsin. Biochemistry. 1984 Jun 5;23(12):2662–2667. doi: 10.1021/bi00307a019. [DOI] [PubMed] [Google Scholar]
- Harbison G. S., Smith S. O., Pardoen J. A., Winkel C., Lugtenburg J., Herzfeld J., Mathies R., Griffin R. G. Dark-adapted bacteriorhodopsin contains 13-cis, 15-syn and all-trans, 15-anti retinal Schiff bases. Proc Natl Acad Sci U S A. 1984 Mar;81(6):1706–1709. doi: 10.1073/pnas.81.6.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms V., Wade R. C. Thermodynamics of water mediating protein-ligand interactions in cytochrome P450cam: a molecular dynamics study. Biophys J. 1995 Sep;69(3):810–824. doi: 10.1016/S0006-3495(95)79955-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R., Baldwin J. M., Ceska T. A., Zemlin F., Beckmann E., Downing K. H. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol. 1990 Jun 20;213(4):899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- Herzfeld J., Das Gupta S. K., Farrar M. R., Harbison G. S., McDermott A. E., Pelletier S. L., Raleigh D. P., Smith S. O., Winkel C., Lugtenburg J. Solid-state 13C NMR study of tyrosine protonation in dark-adapted bacteriorhodopsin. Biochemistry. 1990 Jun 12;29(23):5567–5574. doi: 10.1021/bi00475a022. [DOI] [PubMed] [Google Scholar]
- Holz M., Drachev L. A., Mogi T., Otto H., Kaulen A. D., Heyn M. P., Skulachev V. P., Khorana H. G. Replacement of aspartic acid-96 by asparagine in bacteriorhodopsin slows both the decay of the M intermediate and the associated proton movement. Proc Natl Acad Sci U S A. 1989 Apr;86(7):2167–2171. doi: 10.1073/pnas.86.7.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W., Logunov I., Schulten K., Sheves M. Molecular dynamics study of bacteriorhodopsin and artificial pigments. Biochemistry. 1994 Mar 29;33(12):3668–3678. doi: 10.1021/bi00178a025. [DOI] [PubMed] [Google Scholar]
- Lewis A., Marcus M. A., Ehrenberg B., Crespi H. Experimental evidence for secondary protein-chromophore interactions at the Schiff base linkage in bacteriorhodopsin: Molecular mechanism for proton pumping. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4642–4646. doi: 10.1073/pnas.75.10.4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda A., Sasaki J., Yamazaki Y., Needleman R., Lanyi J. K. Interaction of aspartate-85 with a water molecule and the protonated Schiff base in the L intermediate of bacteriorhodopsin: a Fourier-transform infrared spectroscopic study. Biochemistry. 1994 Feb 22;33(7):1713–1717. doi: 10.1021/bi00173a013. [DOI] [PubMed] [Google Scholar]
- Marti T., Otto H., Mogi T., Rösselet S. J., Heyn M. P., Khorana H. G. Bacteriorhodopsin mutants containing single substitutions of serine or threonine residues are all active in proton translocation. J Biol Chem. 1991 Apr 15;266(11):6919–6927. [PubMed] [Google Scholar]
- Meyer E. Internal water molecules and H-bonding in biological macromolecules: a review of structural features with functional implications. Protein Sci. 1992 Dec;1(12):1543–1562. doi: 10.1002/pro.5560011203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogi T., Stern L. J., Hackett N. R., Khorana H. G. Bacteriorhodopsin mutants containing single tyrosine to phenylalanine substitutions are all active in proton translocation. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5595–5599. doi: 10.1073/pnas.84.16.5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogi T., Stern L. J., Hackett N. R., Khorana H. G. Bacteriorhodopsin mutants containing single tyrosine to phenylalanine substitutions are all active in proton translocation. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5595–5599. doi: 10.1073/pnas.84.16.5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nina M., Roux B., Smith J. C. Functional interactions in bacteriorhodopsin: a theoretical analysis of retinal hydrogen bonding with water. Biophys J. 1995 Jan;68(1):25–39. doi: 10.1016/S0006-3495(95)80184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oesterhelt D., Stoeckenius W. Functions of a new photoreceptor membrane. Proc Natl Acad Sci U S A. 1973 Oct;70(10):2853–2857. doi: 10.1073/pnas.70.10.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oesterhelt D., Stoeckenius W. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat New Biol. 1971 Sep 29;233(39):149–152. doi: 10.1038/newbio233149a0. [DOI] [PubMed] [Google Scholar]
- Otto H., Marti T., Holz M., Mogi T., Lindau M., Khorana H. G., Heyn M. P. Aspartic acid-96 is the internal proton donor in the reprotonation of the Schiff base of bacteriorhodopsin. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9228–9232. doi: 10.1073/pnas.86.23.9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos G., Dencher N. A., Zaccai G., Büldt G. Water molecules and exchangeable hydrogen ions at the active centre of bacteriorhodopsin localized by neutron diffraction. Elements of the proton pathway? J Mol Biol. 1990 Jul 5;214(1):15–19. doi: 10.1016/0022-2836(90)90140-h. [DOI] [PubMed] [Google Scholar]
- Rothschild K. J., He Y. W., Sonar S., Marti T., Khorana H. G. Vibrational spectroscopy of bacteriorhodopsin mutants. Evidence that Thr-46 and Thr-89 form part of a transient network of hydrogen bonds. J Biol Chem. 1992 Jan 25;267(3):1615–1622. [PubMed] [Google Scholar]
- Sampogna R. V., Honig B. Environmental effects on the protonation states of active site residues in bacteriorhodopsin. Biophys J. 1994 May;66(5):1341–1352. doi: 10.1016/S0006-3495(94)80925-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. Y., Mark A. E., Wang C. X., Huang F., Berendsen H. J., van Gunsteren W. F. Can the stability of protein mutants be predicted by free energy calculations? Protein Eng. 1993 Apr;6(3):289–295. doi: 10.1093/protein/6.3.289. [DOI] [PubMed] [Google Scholar]
- Smith S. O., Courtin J., van den Berg E., Winkel C., Lugtenburg J., Herzfeld J., Griffin R. G. Solid-state 13C NMR of the retinal chromophore in photointermediates of bacteriorhodopsin: characterization of two forms of M. Biochemistry. 1989 Jan 10;28(1):237–243. doi: 10.1021/bi00427a033. [DOI] [PubMed] [Google Scholar]
- Song L., El-Sayed M. A., Lanyi J. K. Protein catalysis of the retinal subpicosecond photoisomerization in the primary process of bacteriorhodopsin photosynthesis. Science. 1993 Aug 13;261(5123):891–894. doi: 10.1126/science.261.5123.891. [DOI] [PubMed] [Google Scholar]
- Stern L. J., Khorana H. G. Structure-function studies on bacteriorhodopsin. X. Individual substitutions of arginine residues by glutamine affect chromophore formation, photocycle, and proton translocation. J Biol Chem. 1989 Aug 25;264(24):14202–14208. [PubMed] [Google Scholar]
- Subramaniam S., Marti T., Khorana H. G. Protonation state of Asp (Glu)-85 regulates the purple-to-blue transition in bacteriorhodopsin mutants Arg-82----Ala and Asp-85----Glu: the blue form is inactive in proton translocation. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1013–1017. doi: 10.1073/pnas.87.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade R. C., Mazor M. H., McCammon J. A., Quiocho F. A. A molecular dynamics study of thermodynamic and structural aspects of the hydration of cavities in proteins. Biopolymers. 1991 Jul;31(8):919–931. doi: 10.1002/bip.360310802. [DOI] [PubMed] [Google Scholar]
- Williams M. A., Goodfellow J. M., Thornton J. M. Buried waters and internal cavities in monomeric proteins. Protein Sci. 1994 Aug;3(8):1224–1235. doi: 10.1002/pro.5560030808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccai G. Structure and hydration of purple membranes in different conditions. J Mol Biol. 1987 Apr 5;194(3):569–572. doi: 10.1016/0022-2836(87)90683-8. [DOI] [PubMed] [Google Scholar]
- Zhou F., Windemuth A., Schulten K. Molecular dynamics study of the proton pump cycle of bacteriorhodopsin. Biochemistry. 1993 Mar 9;32(9):2291–2306. doi: 10.1021/bi00060a022. [DOI] [PubMed] [Google Scholar]
- de Groot H. J., Smith S. O., Courtin J., van den Berg E., Winkel C., Lugtenburg J., Griffin R. G., Herzfeld J. Solid-state 13C and 15N NMR study of the low pH forms of bacteriorhodopsin. Biochemistry. 1990 Jul 24;29(29):6873–6883. doi: 10.1021/bi00481a017. [DOI] [PubMed] [Google Scholar]


