Abstract
The classification of white cells plays an important part in medical diagnosis. The counts may suggest the presence of infection, inflammation, anemia, bleeding, and other blood-associated issues. More specifically, the counting in our study is the calculation of the Monocyte Index (MI). The purpose of MI is to determine whether the patient can receive units of blood by analyzing the assay. In case of incompatible blood transfusion, monocytes may ingest or adhere red cells. The index is the percentage of red cells adhered, ingested, or both, versus free monocytes. Manual methods for blood cell counting may take several hours and are highly prone to different sources of errors. Automatic methods, such as Linear Discriminant Analysis, Quadratic Discriminant Analysis, K-Nearest Neighbors, Naïve Bayes, Support Vector Machine, Convolutional Neural Network (CNN), Fast Region-based CNN, Faster Region-based CNN, Spatial Pyramidal Pooling network, Single Shot Detector and Mask Region-based CNN, exist for classification. However, these methods currently do not perform automatic counting and calculation of MI. The dataset is our own collection of images using ZEISS Axiocam 208 color/202 mono microscope camera. For the labels in our own collection, we performed polygonal annotation using the VGG Annotator tool. We trained the Mask R-CNN deep neural network model for automatic segmentation at the pixel-level, using COCO pre-trained weights. Our results look promising, as the Mask R-CNN can perform automatic segmentation with 72% accuracy. Compared to a medical laboratory scientist, the model can process large amount of data simultaneously, quickly and efficiently, with approximately the same judgment accuracy as a human eye. This may significantly reduce the burden of the laboratory scientist and provide a useful reference for doctors to identify a potential blood candidate to be transfused.
Keywords: Blood, Transfusion, Deep Learning, Neural Network, Image Classification
i. Introduction
Image segmentation has been used in the medical field for long time ago. Artificial intelligence algorithms have an incredible ability to learn and improve themselves. The Monocyte Monolayer Assay (MMA) is an in-vitro procedure that has been used for more than 40 years to predict in-vivo survivability of incompatible RBCs [1]. The MMA mimics extravascular hemolysis of red blood cells and may be used to guide transfusion recommendations. The Monocyte Index is a percentage of RBCs adhered, ingested or both by the monocytes. A minimum of 400 myocytes must be counted for enough data to determine the risk of transfusion of incompatible blood. In this work, Artificial Intelligence has been used to automate the Monocyte Index calculation, providing faster results. Mask R-CNN has been implemented in complement to image segmentation and object detection. A COCO pre-trained model was used to fine-tune our own model.
Our key contribution is the first application of deep learning for classification of Monocytes and for Monocyte Index computation. The key significance of this work is that it may potentially save precious time in a life-saving procedure of blood transfusion. The broad significance is that similar methods of segmentation of multiple biological cells may be of great benefit to biological researchers that may want an automated method, as opposed to manual methods of classification and counting. With state-of-the-art deep learning models, we now have more accurate methods in this domain.
A. Background
The classification of white cells, red blood cells and their count have been an important instrument in the Clinical laboratory industry. Gives relevant clinical information about different kinds of disease. Any deviation in the count may suggest the presence of infection, inflammation, anemia, bleeding, and other blood-associated issues [2]. The classification of white cells plays an important part in medical diagnosis, as well. Both are performed by the hematologist by taking a small smear of blood and careful examination under the microscope. Current microscopic methods used for blood cell counting are very tedious and are highly prone to different sources of errors.
MMA is an in-vitro procedure that mimics extravascular hemolysis and can predict the clinical significance of an antibody to guide transfusion recommendations. This procedure has been used for more than 40 years to predict in-vivo survivability of incompatible RBCs. The purpose of MMA is to determine whether the patient can receive random incompatible units rather than rare units. The MI is the percentage of RBCs adhered, ingested, or both (for the total) versus free monocytes [1]. A minimum of 400 monocytes must be counted manually under the microscope. Typically, counting the monocytes takes around one to two hours, depending on the experience of the tech who perform the test. Also, technician’s experience influences the accuracy of the Monocyte Index.
Microscopic image processing is a natural image captured with a camera, telescope, microscope, or other type of optical instrument that displays a continuously varying array of shades and color tones [3]. Eye screening is difficult (Figures 1 and 2). In addition to classical eye screening, image-processing techniques provide greater statistical validity and are used in both cytology and histology: the study of cells and the study of tissues’ microscopic structure [4]. These methods are used widely for clinical laboratories and research.
Figure 2,
Ingested/Phagocytized Monocytes
Deep neural networks provide new methods and layered models that are capable of recognizing high dimensional and complex relationships in a data set. Neuronal networks rely on training data to learn and improve their accuracy over time [5]. A Convolutional Neural Network (CNN) is a kind of deep neural network that works on multiplying filters with images in a systematic manner, that may lead to capturing feature representation and may perform classification tasks in the last layers of the model.
B. Literature Review
Computer-based detection and diagnosis of blood cells tend to avoid human error and reduce the time needed to classify cells. Previous work has performed image processing for classification, identification, and counting applied on red cells and white cells, using machine learning methods. [6, 7, 11, 16, 17, 19, 20]. However, classification of Monocytes has not been performed before, to the best of our knowledge.
While Image classification may seem something natural and relatively easy for humans, but it can become complicated as the image becomes more complex with lot of data and noise. Algorithms can classify images accurately through a pipeline of pre-processing, training, validation and testing.
B.1. Image Classification Algorithms
The literature in Image classification methods is very extensive. In this domain, the most used algorithms are the Principal Component Analysis (PCA), Linear Discriminant Analysis (LDA), Quadratic discriminant analysis (QDA), K-Nearest Neighbour (KNN), Naïve Bayes approach, Support vector machine (SVM), and Artificial neural networks (ANN) [6].
B.2. Object Detection techniques used in Neural Network
Object detection is one of the most well-known areas of deep learning. There are numerous sophisticated techniques (algorithms) used to identify and differentiate objects in an image. Object detection is a computer vision approach that allows a software system to recognize, locate, and trace an object in an image. Object detection distinguishes itself by identifying the type of object and their location-specific coordinates in the image. A bounding box is drawn around the object to indicate its location. The bounding box may or may not be accurate in locating the object’s location. The capacity of a detection algorithm to locate an object within an image determines its performance [7]. The principal design structure in object detections algorithms is the Convolutional Neural Network (CNN). A convolution is the basic process of applying a filter to an input to produce an activation. When the same filter is applied to an input multiple times, a feature map is created, displaying the positions and strength of a recognized feature in an input, such as an image. The capacity of convolutional neural networks to learn a large number of filters in parallel particular to a training dataset under the restrictions of a certain predictive modeling problem, such as image classification, is its unique feature. As a result, extremely precise traits appear on input photographs that can be identified everywhere [8].
B.3. Image Segmentation in Neural Network
Image Segmentation, a fundamental stage of the image recognition system, extracts the items of our interest for additional processing such as description or recognition. An image is a collection of pixels. Picture segmentation is the process of classifying each pixel in an image into a certain category, and therefore could be thought of as a pixel-by-pixel classification problem. There are two types of image segmentation: Sematic segmentation and instance segmentation.
The process of providing a label to each pixel in an image is known as semantic segmentation. Multiple items of the same class are treated as a single unit by semantic segmentation [6]. The purpose of instance segmentation is to provide a view that divides objects of the same class into separate instances [24]. To put it another way, semantic segmentation treats several objects belonging to the same category as a single entity. On the other hand, instance segmentation recognizes specific objects within these categories.
In deep learning there is a variety of methods used in image segmentation as Fully convolutional Network [9], U-net [10], Deep-Lab [11] Global Convolution Network [12], Tiramisu Model [13], Hybrid CNN-CRF methods [14], and See More than Once – KSAC [15], among others.
B.4. Relationship between object detection, classification, and segmentation.
It’s important to differentiate between each category to understand better how they work together. The classification consists of developing a prediction for the entire input. The next phase is detection, which not only provides the classes but also extra information about their specific position. Finally, semantic segmentation achieves fine-grained inference by making dense predictions inferring labels for each pixel, resulting in each pixel being labeled with the class of the object or region it surrounds [25].
B.5. Related literature review
Faster R-CNN and Feature Pyramid Network has been used to differentiate and count white blood cell (WBC) in bone marrow from Leukemia patients, having 98.8% accuracy [16]. Also, Mask – R-CNN has been used to classify and count RBC and WBC by Dhieb et al [17], Mask R-CNN as well has been adapted to detect abnormal RBCs to help in the diagnosis of blood disorders in short period of time [18]. In addition, R-CNN and YOLO has been combined to identify and differentiate white blood cells subtypes, with 99.2% accuracy on detection and 90% accuracy in the classification [19]. Furthermore, Retina Net deep learning network which is used to recognize and classify the main three types of blood cells: erythrocytes, leukocytes and platelets [20].
This work has been designed to help the laboratory technicians to calculate the Monocyte Index (MI) using microscopic images. This task can be challenging based on the degree of difficulty in differentiating the overlapped red blood cells that have been Ingested/Phagocytized, Adhered, and Free Monocytes.
B.6. Methods
We used Mask R-CNN techniques for instance segmentation which allows not only the detection and location of cells but also their classification and counting.
C. Datasets Acquisition and Processing
We use COCO pre-trained weights to train our own model. Also, we use our own dataset collection image collected using ZEISS Axiocam 208 color/202 mono Microscope camera. A total of 500 image were divided in three folders (train, validation, and test).
D. Proposed Solution
Mask-RCNN model has been modified to identify and count the monocytes present in the dataset. A minimum of 400 monocytes it must be identified in order to provide an acceptable Monocyte Index (MI). The formula to calculate the MI is as follows:
The MI is the percentage of RBCs adhered, ingested or both (for the total) verses free monocytes. An MI of ‘Zero’ or 0 indicates there were no adhered or phagocytized red cells [1]. Experience with this procedure has been similar to others; in that MI values of ≤5% have indicated that incompatible blood can be given without the risk of an overt hemolytic transfusion reaction but it does not guarantee normal long-term survival of those RBCs. MI values ranging from 5–20% have a reduced risk of clinical significance, but signs and symptoms of transfusion reaction may occur. Similarly, an MI of >20% indicates the antibody has clinical significance, which may range from abnormal RBC survival to clinically obvious adverse reactions.
E. Results
The neural network has two primary layers: coco transfer learning to speed up computation and enhance accuracy, and object identification and categorization, which supports the preceding layer. A Matterport Mask RCNN was used to identify objects. In the classification layer, a ResNet101 was used. Our training (fine-tuning) model took approximately 8 hours to complete the training with 20 epochs and 500 steps per epoch. Our GPU was a simple AMD Radeon R7 250. We had good quality (uncompressed) images.
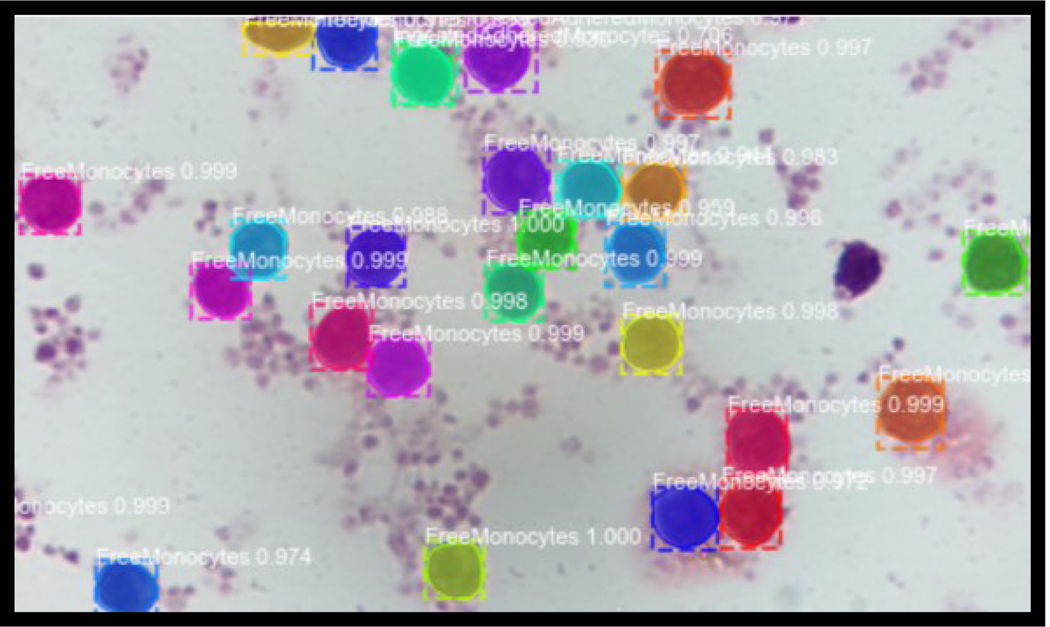
Our model was able to successfully detect, classify and count the two different classes assigned and the background (Figure 3). We used minimum confidence of 0.9, one GPU per image, and NMS threshold of 0.3, as our key parameters. The Confusion Matrix for the Test data is shown at Table 1.0.
Table 1.0.
Confusion Matrix
| True | False | Total | |
|---|---|---|---|
| Positive | 108 | 20 | 128 |
| Negative | 0 | 22 | 22 |
| Total | 108 | 42 | 150 |
The time taken for detection, classification and printing the MI was about 5s. The false positives may be coming from the platelets, the noise in the image and the calibration of the microscope. A visual inspection was performed by a trained laboratory scientist with the results of our model. Compared to the trained laboratory scientist, the model can process with comparable accuracy. Human error can be as high as 30% at times of heavy workload and noisy images.
ii. Summary, Conclusion and Recommendations
Compared to a medical laboratory scientist, the model can process large amounts of data simultaneously, quickly and efficiently, with approximately the same judgment accuracy as a human eye. The laboratory scientist can use the tool for the easy segmentations at high speed. For the uncertain segmentations, the manual methods may also be used for confirmation, which may significantly reduce the burden of the laboratory scientist and provide a useful reference for doctors to identify a potential blood candidate to be transfused. We hope that this method will increase the range of potential approaches to use a real time video analysis of Monocyte Index calculation.
Figure 1,
Free Monocytes
Figure 1,
Example of Monocytes detected and classified
Contributor Information
Luis A. Pena Marquez, Computer Science Louisiana State University Shreveport Shreveport, USA.
Subhajit Chakrabarty, Computer Science Louisiana State University Shreveport Shreveport, USA.
References
- [1].Arndt PA, & Garratty G. (2004). A retrospective analysis of the value of monocyte monolayer assay results for predicting the clinical significance of blood group alloantibodies. Transfusion, 44(9):1273–81. [DOI] [PubMed] [Google Scholar]
- [2].DM H. (2019). Modern blood banking and Transfusion Practices. 7th ed. Philadelphia, PA: F.A. Davis Company. [Google Scholar]
- [3].Spring KR, Russ JC, & Davidson MW (2022, March 14). Basic Concepts in Digital Image Processing. From OLYMPUS: https://www.olympus-lifescience.com/es/microscope-resource/primer/digitalimaging/imageprocessingintro/ [Google Scholar]
- [4].Smochina C, Herghelegiu P, & Manta V. (2011). Image Processing Techniques used in Microscopic Image Segmentation. Faculty of Automatic Control and Computer Engineering, 84–98. From Faculty of Automatic Control and Computer Engineering. [Google Scholar]
- [5].Chen J. (2021, December 08). Neural Network. Retrieved from Investopedia: https://www.investopedia.com/terms/n/neuralnetwork.asp#:~:text=A%20neural%20network%20is%20a,organic%20or%20artificial%20in%20nature [Google Scholar]
- [6].Nufer G, & Muth M. (2022). Artificial Intelligence in Marketing Analytics: The Application of Artificial Neural Networks for Brand Image Measurement. Journal of Marketing Development and Competitiveness, 16(2022), 56. From https://eds.p.ebscohost.com/eds/pdfviewer/pdfviewer?vid=3&sid=b2c86439-f180-4d66-9758-c8a6c55052d1%40redis [Google Scholar]
- [7].Team GL (2021, December 25). Real-Time Object Detection Using TensorFlow. From Great Leearning: https://www.mygreatlearning.com/blog/object-detection-using-tensorflow/#sh1 [Google Scholar]
- [8].Brownlee J. (2020, April 17). How Do Convolutional Layers Work in Deep Learning Neural Networks? From Machine Learning Mastery: https://machinelearningmastery.com/convolutional-layers-for-deep-learning-neural-networks/ [Google Scholar]
- [9].Long J, Shelhamer E, & Darrel T. (2015). Fully Convolutional Networks for Semantic Segmentation. IEEE Xplore, 1–10. [DOI] [PubMed] [Google Scholar]
- [10].Ronneberger O, Fischer P, & Brox T. (2015). U-Net: Convolutional Networks for Biomedical. VISION, 1–8. [Google Scholar]
- [11].Chen L-C, Papandreou G, Kokkinos I, Murphy K, & Yuille A. (2017). DeepLab: Semantic Image Segmentation with Deep Convolutional Nets, Atrous Convolution, and Fully Connected CRFs. ARXIV, 1–14. [DOI] [PubMed] [Google Scholar]
- [12].Peng C, Zhang X, Yu G, Luo G, & Sun J. (2017). Large Kernel Matters - Improve Semantic Segmentation by Global Convolutional Network. IEEE Xplore, 1–9. [Google Scholar]
- [13].Jgou S, Drozdzal M, Vazquez D, Romero D, Romero A, & Bengio Y. (2017). The One Hundred Layers Tiramisu: Fully Convolutional DenseNets for Semantic Segmentation. ARXIV, 1–9. [Google Scholar]
- [14].Zheng S, Jayasumana S, Romera-Paredes B, Vineet V, Su Z, Du D, . . . Tsorr PH (2015). The One Hundred Layers Tiramisu:. University of Oxford, 1–17. [Google Scholar]
- [15].Anil Chandra NM (2021, May 21). A 2021 guide to Semantic Segmentation. From Nanonets: https://nanonets.com/blog/semantic-image-segmentation-2020/ [Google Scholar]
- [16].Wang D, Hwang M, Jiang W-C, Ding K, Chang HC, & Hwang K-S (2021). A deep learning method for counting white blood cells in bone marrow images. BMC Bioinformatics, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Dhieb N, Ghazzai H, & Besbes H. (2019). An Automated Blood Cells Counting and Classification Framework using Mask R-CNN Deep Learning Model. ResearchGate, 1–6. [Google Scholar]
- [18].Lin Y-H, Liao K. Y. k., & Sung K-B (2020). Automatic detection and characterization of quantitative phase images of thalassemic red blood cells using a mask region-based convolutional neural network. Journal of Biomedical Optics, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Praveen N, Punn NS, Sonbhadra SK, Agarwal S, Syafrullah M, & Adiyarta K. (2021). White blood cell subtype detection and classification. ARXIV, 1–5. [Google Scholar]
- [20].Dralus G, Mazur D, & Czmil A. (n.d.). Automatic Detection and Counting of Blood Cells in Smear Images Using RetinaNet. [DOI] [PMC free article] [PubMed] [Google Scholar]