Abstract
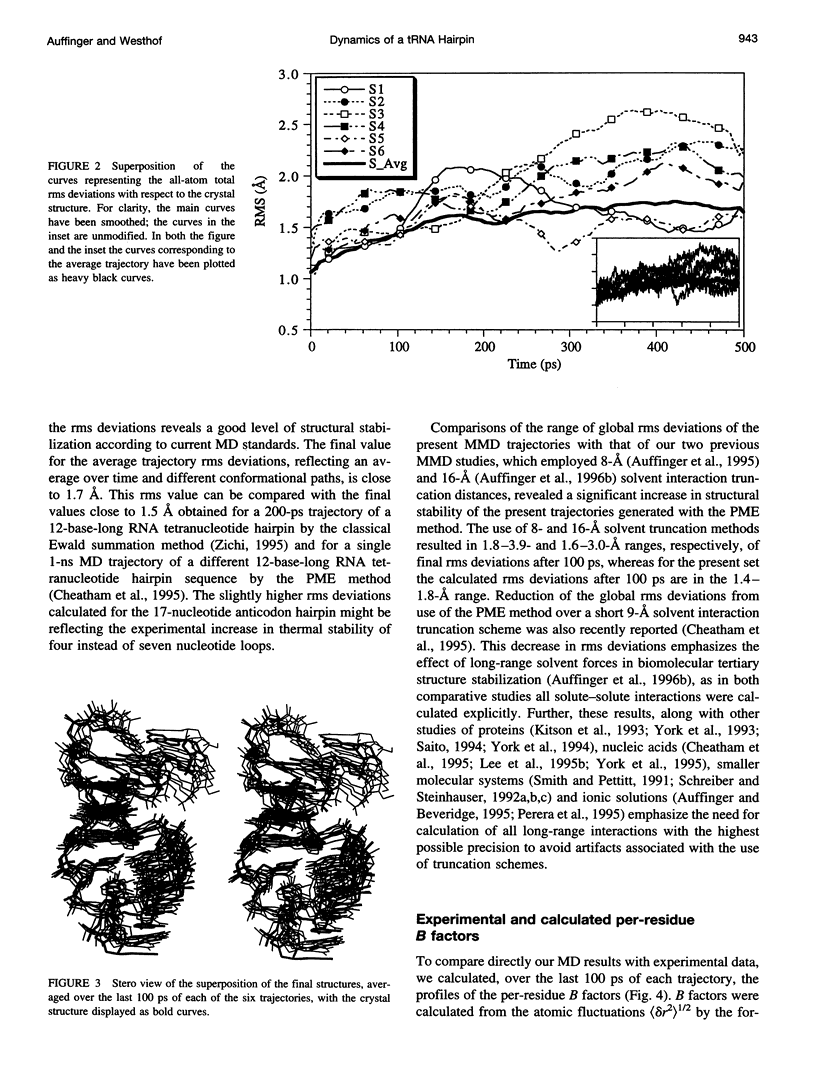
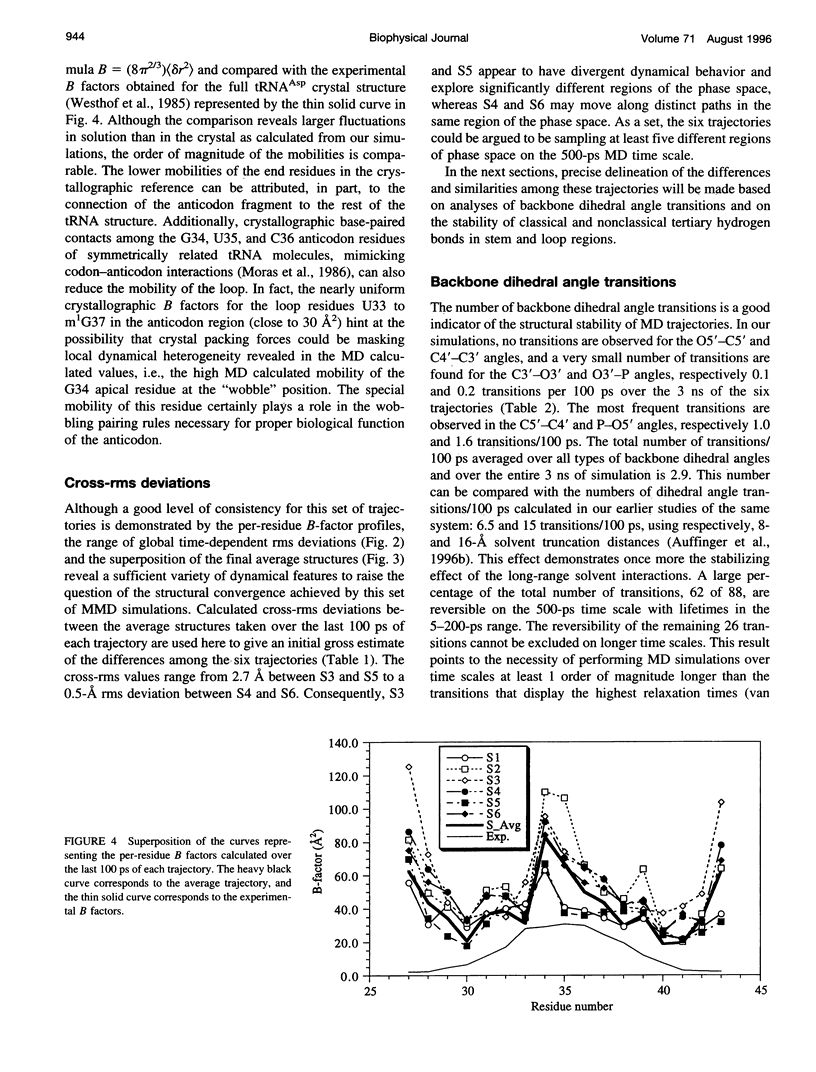
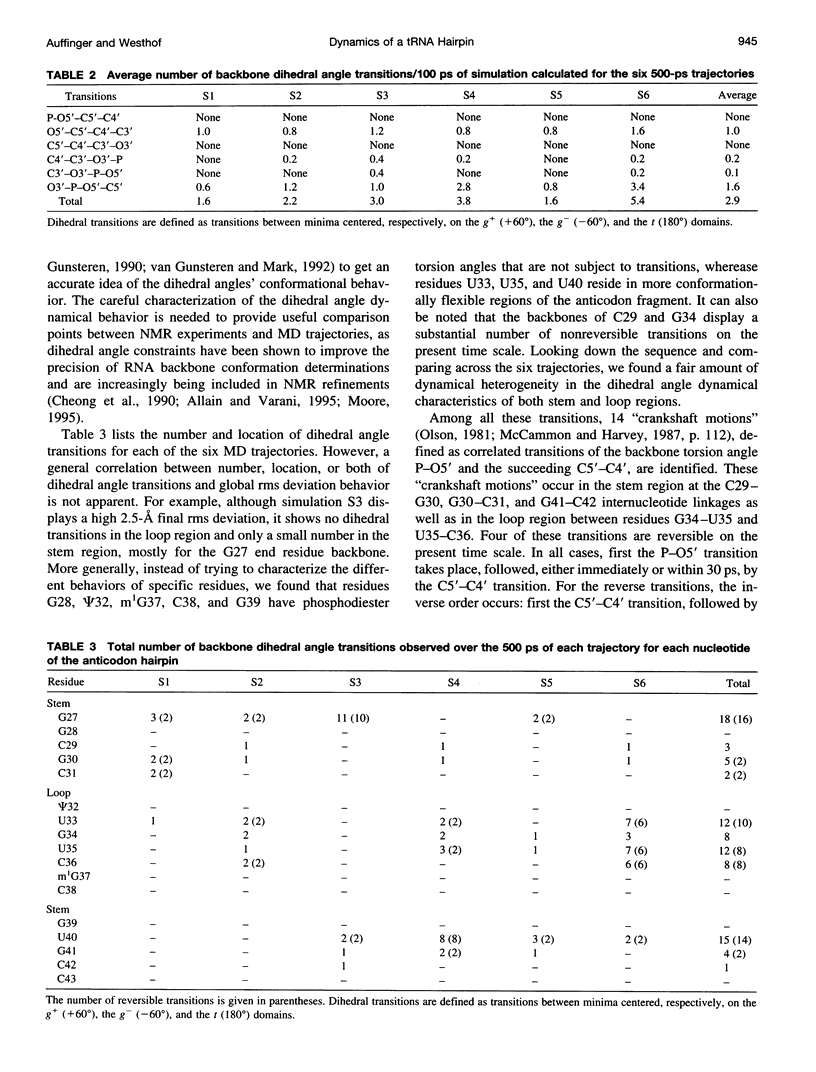
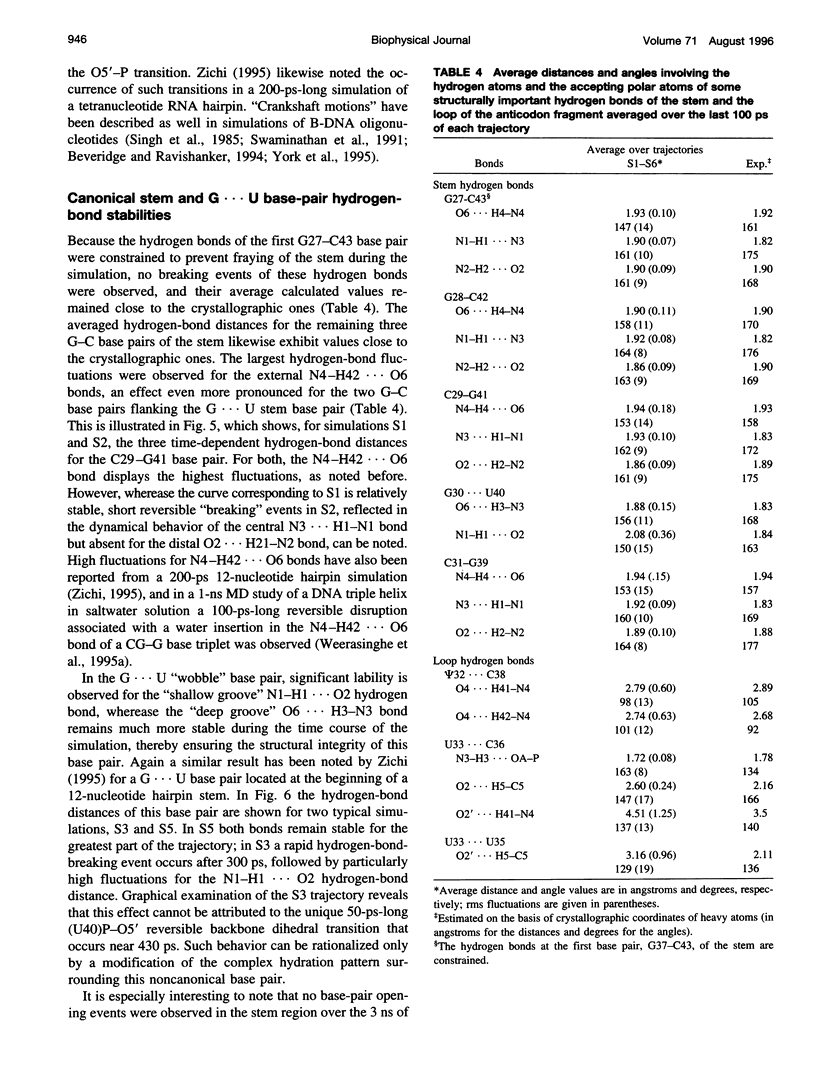
Multiple molecular dynamics trajectories of the solvated and neutralized 17-residue tRNA(Asp) anticodon hairpin were generated for a total of 3 ns. Explicit treatment of all long-ranged electrostatic interactions by the particle mesh Ewald algorithm, as implemented in the AMBER MD software package, effected a degree of structural stabilization not previously achieved by use of a long 16-A solvent interaction truncation scheme. The increased stability of this multiple molecular dynamics set was appropriate for an in-depth analysis of the six 500-ps-long trajectories and allowed the characterization of a number of key structural interactions. The dynamical behavior of the standard Watson-Crick base pairs, the noncanonical G30-U40 "wobble" base pair, and the psi 32-C38 pseudo-base pair is presented as well as that of two C--H... O hydrogen bonds found to contribute to the array of tertiary interactions that stabilize the seven-nucleotide native loop conformation. The least mobile residue in the loop is U33, which forms the U-turn motif and which participates in several hydrogen-bonding interactions, whereas the most mobile residue is the apical residue G34 at the wobble position, a factor undoubtedly important in its biological function. The set of multiple molecular dynamics trajectories obtained does not converge on a 500-ps time scale to a unique dynamical model but instead describes an ensemble of structural microstates accessible to the system under the present simulation protocol, which is the result of local structural heterogeneity rather than of global conformational changes.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allain F. H., Varani G. Structure of the P1 helix from group I self-splicing introns. J Mol Biol. 1995 Jul 14;250(3):333–353. doi: 10.1006/jmbi.1995.0381. [DOI] [PubMed] [Google Scholar]
- Berman H. M., Olson W. K., Beveridge D. L., Westbrook J., Gelbin A., Demeny T., Hsieh S. H., Srinivasan A. R., Schneider B. The nucleic acid database. A comprehensive relational database of three-dimensional structures of nucleic acids. Biophys J. 1992 Sep;63(3):751–759. doi: 10.1016/S0006-3495(92)81649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R. S., Dewan J. C., Klug A. Crystallographic and biochemical investigation of the lead(II)-catalyzed hydrolysis of yeast phenylalanine tRNA. Biochemistry. 1985 Aug 27;24(18):4785–4801. doi: 10.1021/bi00339a012. [DOI] [PubMed] [Google Scholar]
- Cheong C., Varani G., Tinoco I., Jr Solution structure of an unusually stable RNA hairpin, 5'GGAC(UUCG)GUCC. Nature. 1990 Aug 16;346(6285):680–682. doi: 10.1038/346680a0. [DOI] [PubMed] [Google Scholar]
- Claesson C., Lustig F., Borén T., Simonsson C., Barciszewska M., Lagerkvist U. Glycine codon discrimination and the nucleotide in position 32 of the anticodon loop. J Mol Biol. 1995 Mar 24;247(2):191–196. doi: 10.1006/jmbi.1994.0132. [DOI] [PubMed] [Google Scholar]
- Derewenda Z. S., Lee L., Derewenda U. The occurrence of C-H...O hydrogen bonds in proteins. J Mol Biol. 1995 Sep 15;252(2):248–262. doi: 10.1006/jmbi.1995.0492. [DOI] [PubMed] [Google Scholar]
- Kitson D. H., Avbelj F., Moult J., Nguyen D. T., Mertz J. E., Hadzi D., Hagler A. T. On achieving better than 1-A accuracy in a simulation of a large protein: Streptomyces griseus protease A. Proc Natl Acad Sci U S A. 1993 Oct 1;90(19):8920–8924. doi: 10.1073/pnas.90.19.8920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochoyan M., Leroy J. L., Guéron M. Processes of base-pair opening and proton exchange in Z-DNA. Biochemistry. 1990 May 22;29(20):4799–4805. doi: 10.1021/bi00472a008. [DOI] [PubMed] [Google Scholar]
- Leonard G. A., McAuley-Hecht K., Brown T., Hunter W. N. Do C-H...O hydrogen bonds contribute to the stability of nucleic acid base pairs? Acta Crystallogr D Biol Crystallogr. 1995 Mar 1;51(Pt 2):136–139. doi: 10.1107/S0907444994004713. [DOI] [PubMed] [Google Scholar]
- Louise-May S., Auffinger P., Westhof E. Calculations of nucleic acid conformations. Curr Opin Struct Biol. 1996 Jun;6(3):289–298. doi: 10.1016/s0959-440x(96)80046-7. [DOI] [PubMed] [Google Scholar]
- Moe J. G., Russu I. M. Proton exchange and base-pair opening kinetics in 5'-d(CGCGAATTCGCG)-3' and related dodecamers. Nucleic Acids Res. 1990 Feb 25;18(4):821–827. doi: 10.1093/nar/18.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moras D., Dock A. C., Dumas P., Westhof E., Romby P., Ebel J. P., Giegé R. Anticodon-anticodon interaction induces conformational changes in tRNA: yeast tRNAAsp, a model for tRNA-mRNA recognition. Proc Natl Acad Sci U S A. 1986 Feb;83(4):932–936. doi: 10.1073/pnas.83.4.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearlman D. A., Kim S. H. Atomic charges for DNA constituents derived from single-crystal X-ray diffraction data. J Mol Biol. 1990 Jan 5;211(1):171–187. doi: 10.1016/0022-2836(90)90019-I. [DOI] [PubMed] [Google Scholar]
- Pley H. W., Flaherty K. M., McKay D. B. Three-dimensional structure of a hammerhead ribozyme. Nature. 1994 Nov 3;372(6501):68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]
- Quigley G. J., Rich A. Structural domains of transfer RNA molecules. Science. 1976 Nov 19;194(4267):796–806. doi: 10.1126/science.790568. [DOI] [PubMed] [Google Scholar]
- Ruff M., Krishnaswamy S., Boeglin M., Poterszman A., Mitschler A., Podjarny A., Rees B., Thierry J. C., Moras D. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp). Science. 1991 Jun 21;252(5013):1682–1689. doi: 10.1126/science.2047877. [DOI] [PubMed] [Google Scholar]
- Schreiber H., Steinhauser O. Cutoff size does strongly influence molecular dynamics results on solvated polypeptides. Biochemistry. 1992 Jun 30;31(25):5856–5860. doi: 10.1021/bi00140a022. [DOI] [PubMed] [Google Scholar]
- Schreiber H., Steinhauser O. Taming cut-off induced artifacts in molecular dynamics studies of solvated polypeptides. The reaction field method. J Mol Biol. 1992 Dec 5;228(3):909–923. doi: 10.1016/0022-2836(92)90874-j. [DOI] [PubMed] [Google Scholar]
- Scott W. G., Finch J. T., Klug A. The crystal structure of an all-RNA hammerhead ribozyme: a proposed mechanism for RNA catalytic cleavage. Cell. 1995 Jun 30;81(7):991–1002. doi: 10.1016/s0092-8674(05)80004-2. [DOI] [PubMed] [Google Scholar]
- Singh U. C., Weiner S. J., Kollman P. Molecular dynamics simulations of d(C-G-C-G-A) X d(T-C-G-C-G) with and without "hydrated" counterions. Proc Natl Acad Sci U S A. 1985 Feb;82(3):755–759. doi: 10.1073/pnas.82.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl M. C., Rao S. T., Sundaralingam M. The structure of r(UUCGCG) has a 5'-UU-overhang exhibiting Hoogsteen-like trans U.U base pairs. Nat Struct Biol. 1996 Jan;3(1):24–31. doi: 10.1038/nsb0196-24. [DOI] [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol. 1985 Jul 5;184(1):119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- York D. M., Wlodawer A., Pedersen L. G., Darden T. A. Atomic-level accuracy in simulations of large protein crystals. Proc Natl Acad Sci U S A. 1994 Aug 30;91(18):8715–8718. doi: 10.1073/pnas.91.18.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gunsteren W. F., Mark A. E. On the interpretation of biochemical data by molecular dynamics computer simulation. Eur J Biochem. 1992 Mar 15;204(3):947–961. doi: 10.1111/j.1432-1033.1992.tb16716.x. [DOI] [PubMed] [Google Scholar]



