Abstract
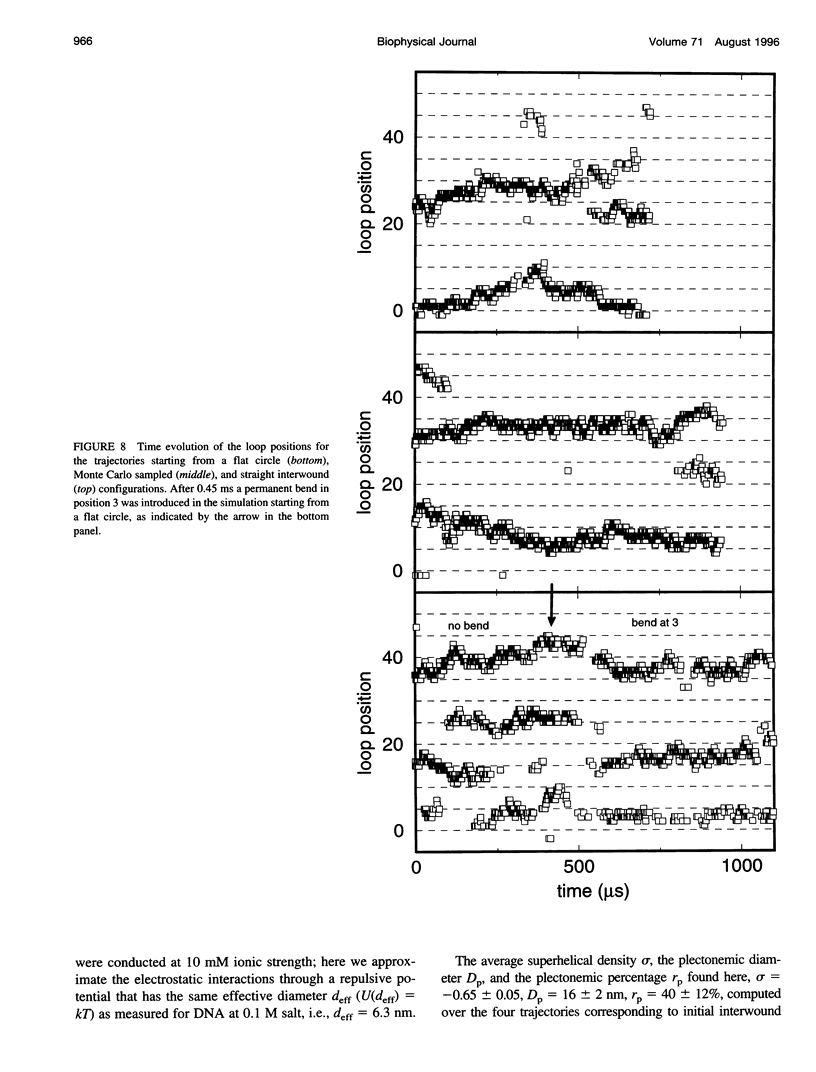
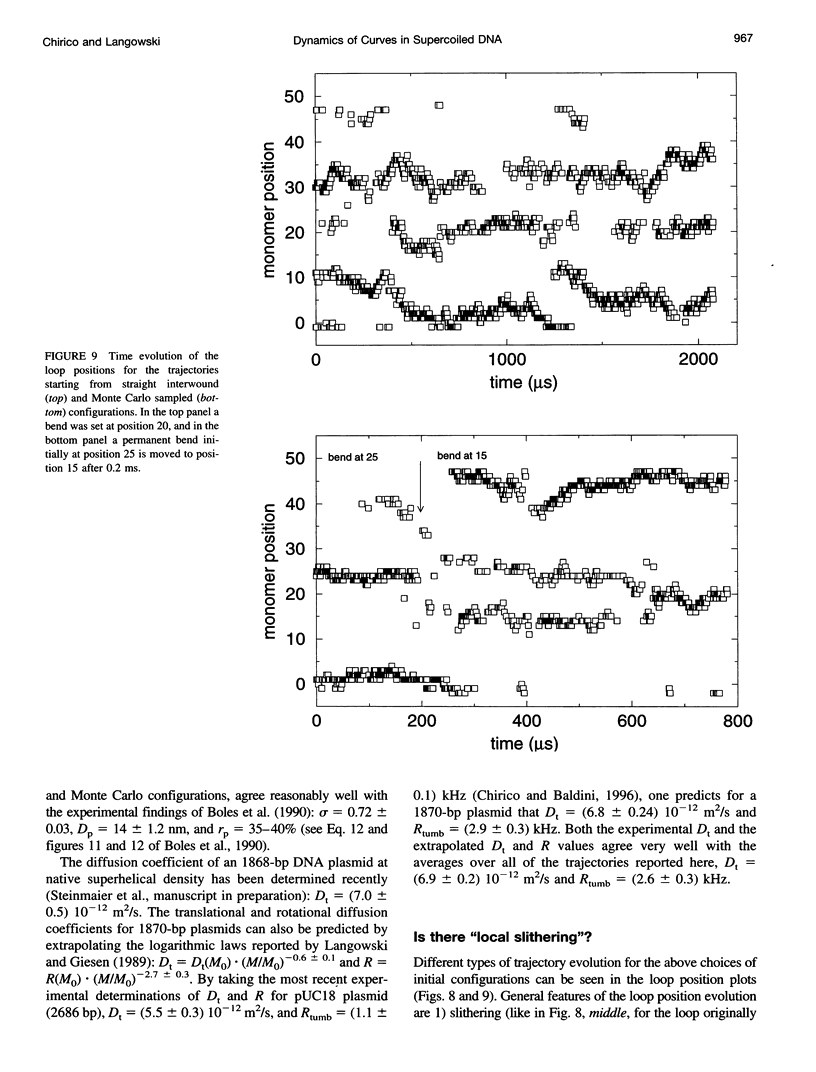
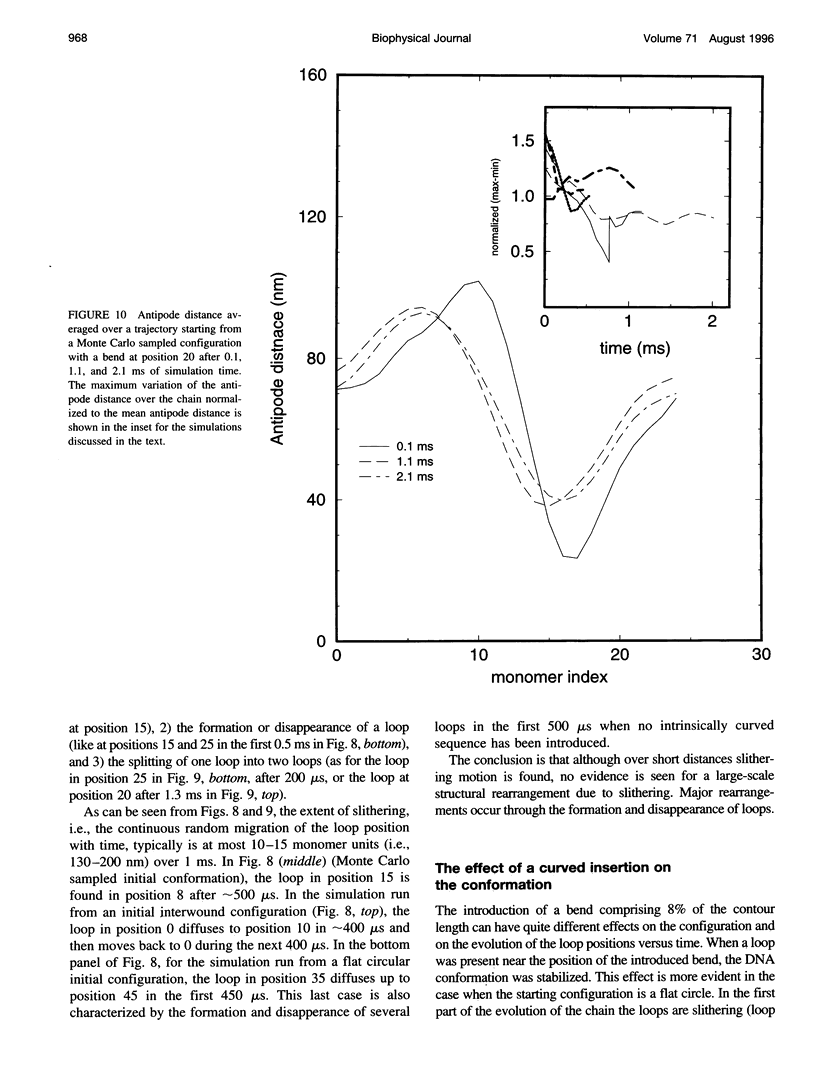
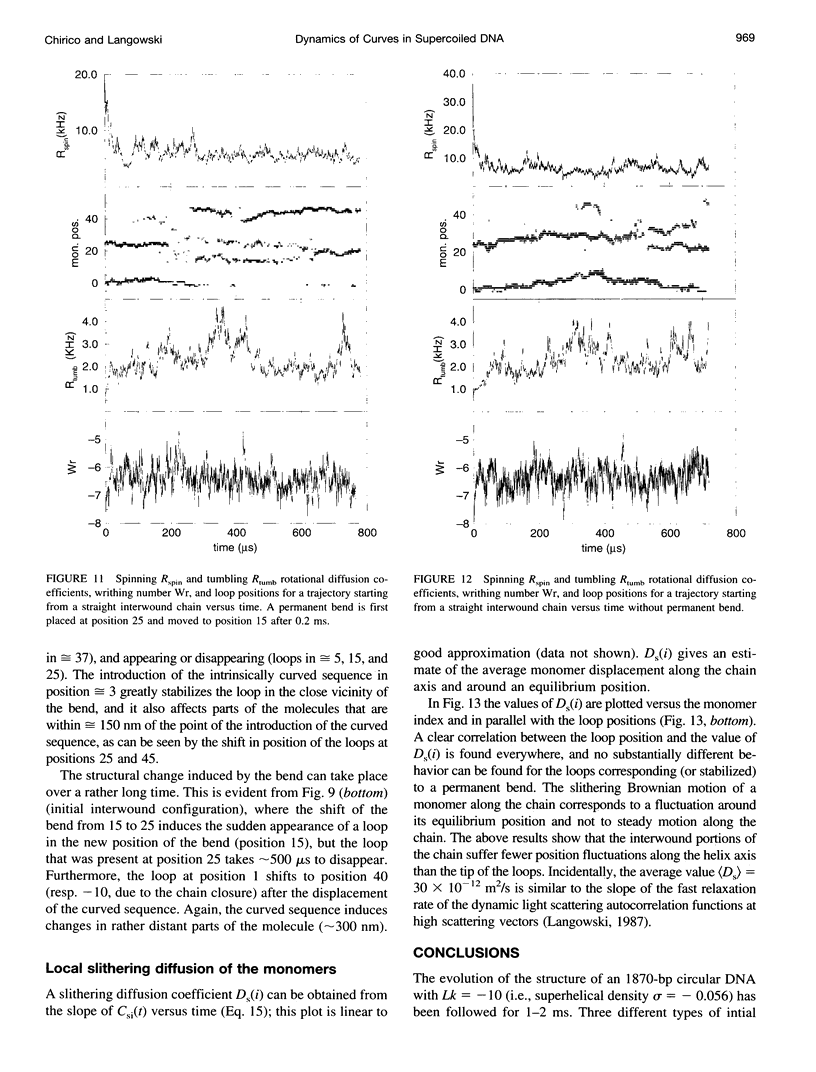
The recently presented Brownian dynamics model for superhelical DNA is extended to include local curvature of the DNA helix axis. Here we analyze the effect of a permanent bend on the structure and dynamics of an 1870-bp superhelix with delta Lk = -10. Furthermore, we define quantitative expressions for computing structural parameters such as loop positions, superhelix diameter, and plectonemic content for trajectories of superhelical DNA, and assess the convergence toward global equilibrium. The structural fluctuations in an interwound superhelix, as reflected in the change in end loop positions, seem to occur by destruction/creation of loops rather than by a sliding motion of the DNA around its contour. Their time scale is on the order of 30-100 microseconds. A permanent bend changes the structure and the internal motions of the DNA drastically. The position of the end loop is fixed at the permanent bend, and the local motions of the chain are enhanced near the loops. A displacement of the bend from the end loop to a position inside the plectonemic part of the superhelix results in the formation of a new loop and the disappearance of the old one; we estimate the time involved in this process to be about 0.5 ms.
Full text
PDF




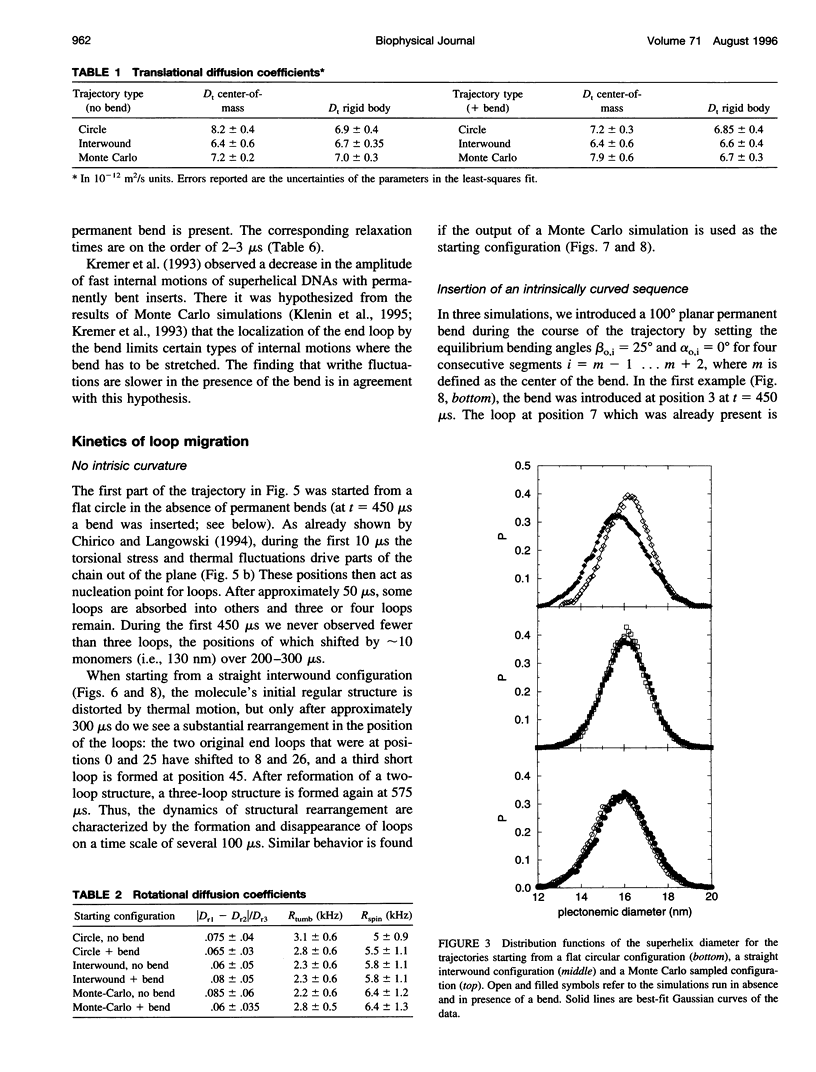
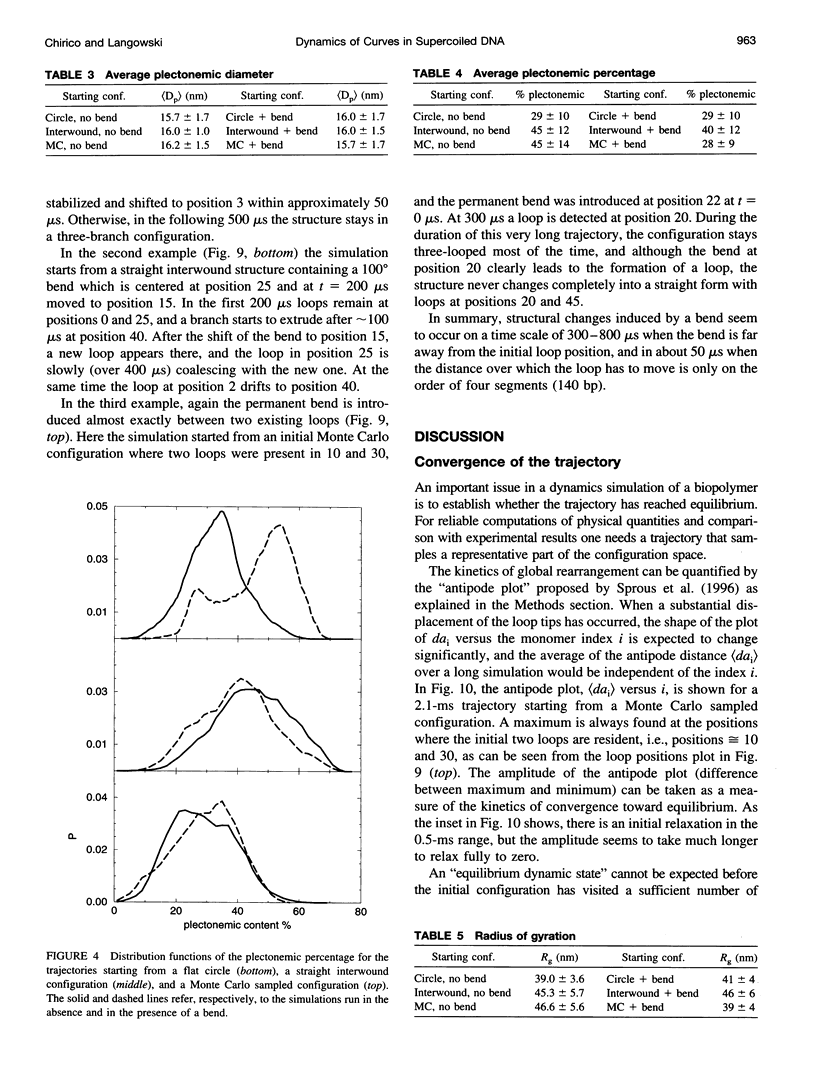
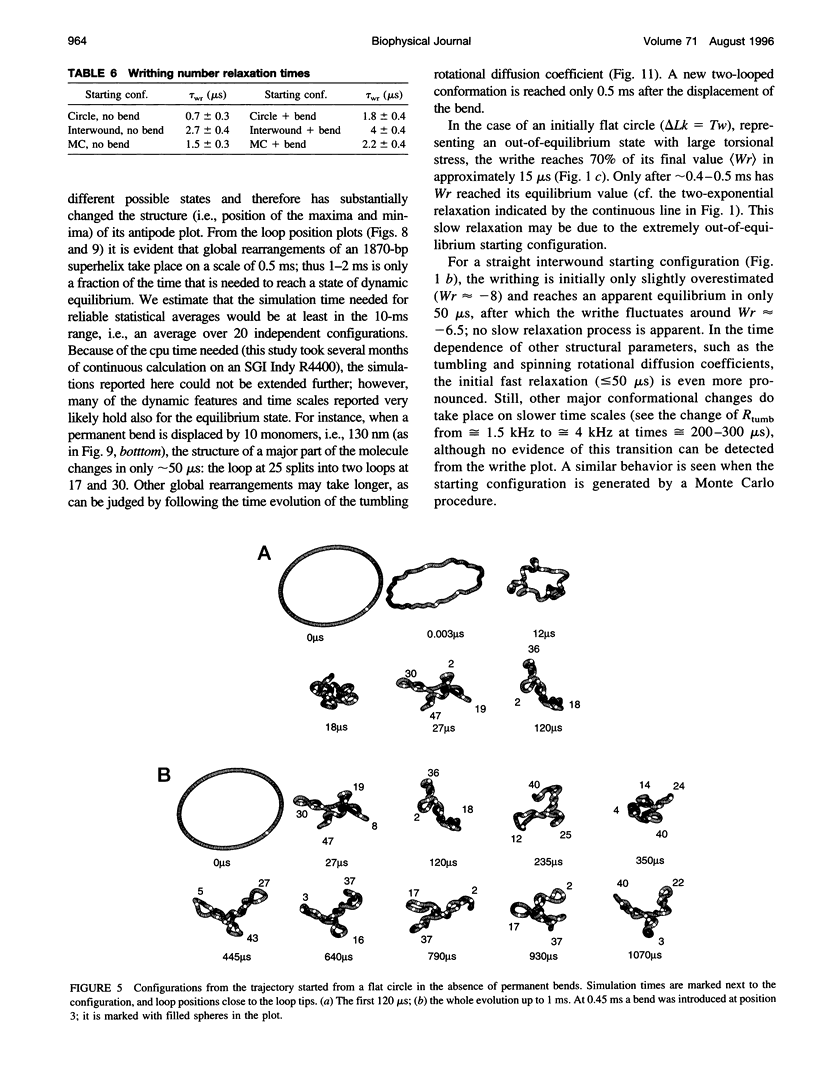
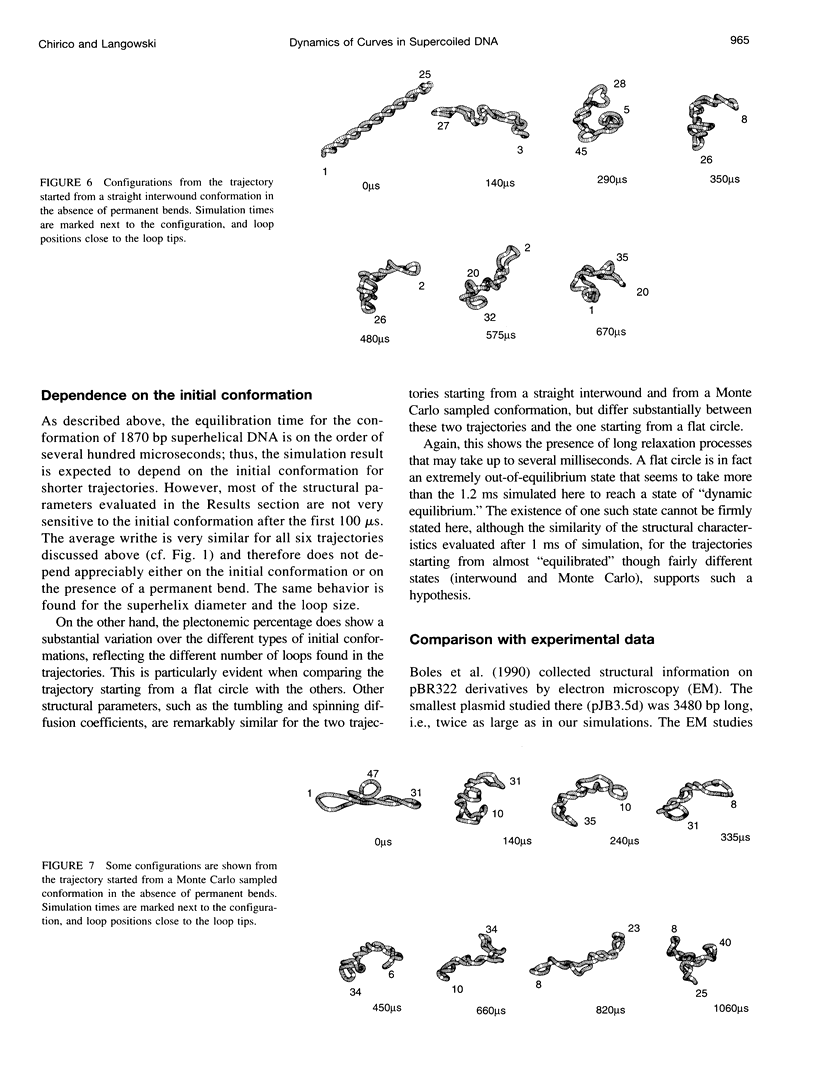

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bednar J., Furrer P., Stasiak A., Dubochet J., Egelman E. H., Bates A. D. The twist, writhe and overall shape of supercoiled DNA change during counterion-induced transition from a loosely to a tightly interwound superhelix. Possible implications for DNA structure in vivo. J Mol Biol. 1994 Jan 21;235(3):825–847. doi: 10.1006/jmbi.1994.1042. [DOI] [PubMed] [Google Scholar]
- Boles T. C., White J. H., Cozzarelli N. R. Structure of plectonemically supercoiled DNA. J Mol Biol. 1990 Jun 20;213(4):931–951. doi: 10.1016/S0022-2836(05)80272-4. [DOI] [PubMed] [Google Scholar]
- Borowiec J. A., Zhang L., Sasse-Dwight S., Gralla J. D. DNA supercoiling promotes formation of a bent repression loop in lac DNA. J Mol Biol. 1987 Jul 5;196(1):101–111. doi: 10.1016/0022-2836(87)90513-4. [DOI] [PubMed] [Google Scholar]
- Collini M., Chirico G., Baldini G., Bianchi M. E. Conformation of short DNA fragments by modulated fluorescence polarization anisotropy. Biopolymers. 1995 Aug;36(2):211–225. doi: 10.1002/bip.360360209. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J. Flexibility of DNA. Annu Rev Biophys Biophys Chem. 1988;17:265–286. doi: 10.1146/annurev.bb.17.060188.001405. [DOI] [PubMed] [Google Scholar]
- Klenin K. V., Frank-Kamenetskii M. D., Langowski J. Modulation of intramolecular interactions in superhelical DNA by curved sequences: a Monte Carlo simulation study. Biophys J. 1995 Jan;68(1):81–88. doi: 10.1016/S0006-3495(95)80161-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer W., Klenin K., Diekmann S., Langowski J. DNA curvature influences the internal motions of supercoiled DNA. EMBO J. 1993 Nov;12(11):4407–4412. doi: 10.1002/j.1460-2075.1993.tb06125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langowski J., Giesen U. Configurational and dynamic properties of different length superhelical DNAs measured by dynamic light scattering. Biophys Chem. 1989 Sep 15;34(1):9–18. doi: 10.1016/0301-4622(89)80036-5. [DOI] [PubMed] [Google Scholar]
- Langowski J., Olson W. K., Pedersen S. C., Tobias I., Westcott T. P., Yang Y. DNA supercoiling, localized bending and thermal fluctuations. Trends Biochem Sci. 1996 Feb;21(2):50–50. [PubMed] [Google Scholar]
- Langowski J. Salt effects on internal motions of superhelical and linear pUC8 DNA. Dynamic light scattering studies. Biophys Chem. 1987 Sep;27(3):263–271. doi: 10.1016/0301-4622(87)80066-2. [DOI] [PubMed] [Google Scholar]
- Laundon C. H., Griffith J. D. Curved helix segments can uniquely orient the topology of supertwisted DNA. Cell. 1988 Feb 26;52(4):545–549. doi: 10.1016/0092-8674(88)90467-9. [DOI] [PubMed] [Google Scholar]
- Rybenkov V. V., Cozzarelli N. R., Vologodskii A. V. Probability of DNA knotting and the effective diameter of the DNA double helix. Proc Natl Acad Sci U S A. 1993 Jun 1;90(11):5307–5311. doi: 10.1073/pnas.90.11.5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlick T. Modeling superhelical DNA: recent analytical and dynamic approaches. Curr Opin Struct Biol. 1995 Apr;5(2):245–262. doi: 10.1016/0959-440x(95)80083-2. [DOI] [PubMed] [Google Scholar]
- Stigter D. Interactions of highly charged colloidal cylinders with applications to double-stranded. Biopolymers. 1977 Jul;16(7):1435–1448. doi: 10.1002/bip.1977.360160705. [DOI] [PubMed] [Google Scholar]
- Vologodskii A. V., Levene S. D., Klenin K. V., Frank-Kamenetskii M., Cozzarelli N. R. Conformational and thermodynamic properties of supercoiled DNA. J Mol Biol. 1992 Oct 20;227(4):1224–1243. doi: 10.1016/0022-2836(92)90533-p. [DOI] [PubMed] [Google Scholar]
- Yang Y., Westcott T. P., Pedersen S. C., Tobias I., Olson W. K. Effects of localized bending on DNA supercoiling. Trends Biochem Sci. 1995 Aug;20(8):313–319. doi: 10.1016/s0968-0004(00)89058-1. [DOI] [PubMed] [Google Scholar]
- Zhang P., Tobias I., Olson W. K. Computer simulation of protein-induced structural changes in closed circular DNA. J Mol Biol. 1994 Sep 23;242(3):271–290. doi: 10.1006/jmbi.1994.1578. [DOI] [PubMed] [Google Scholar]
- ten Heggeler-Bordier B., Wahli W., Adrian M., Stasiak A., Dubochet J. The apical localization of transcribing RNA polymerases on supercoiled DNA prevents their rotation around the template. EMBO J. 1992 Feb;11(2):667–672. doi: 10.1002/j.1460-2075.1992.tb05098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]