Abstract
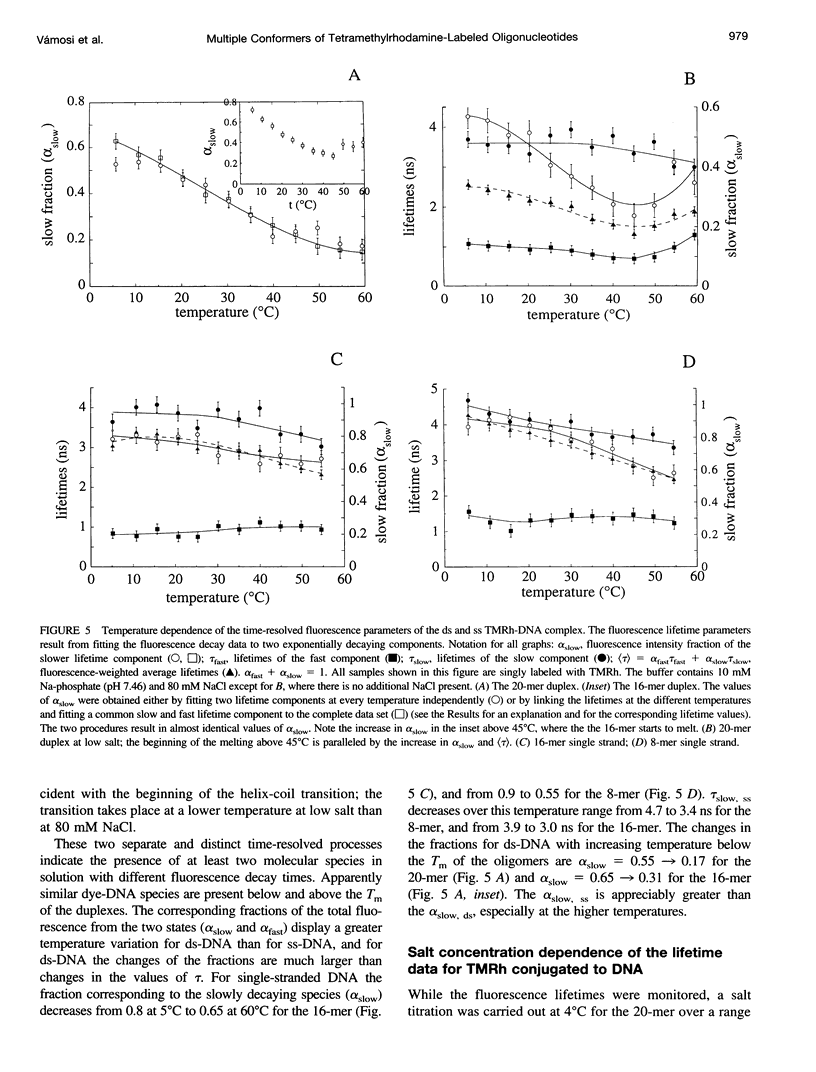
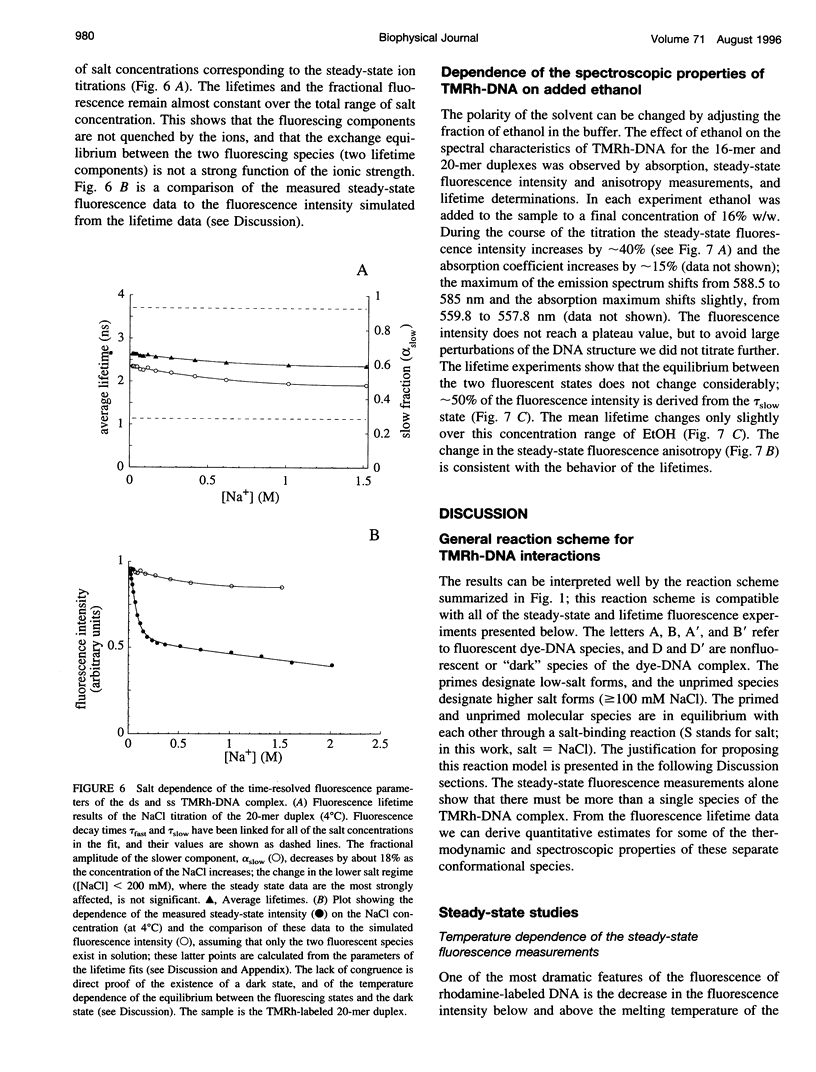
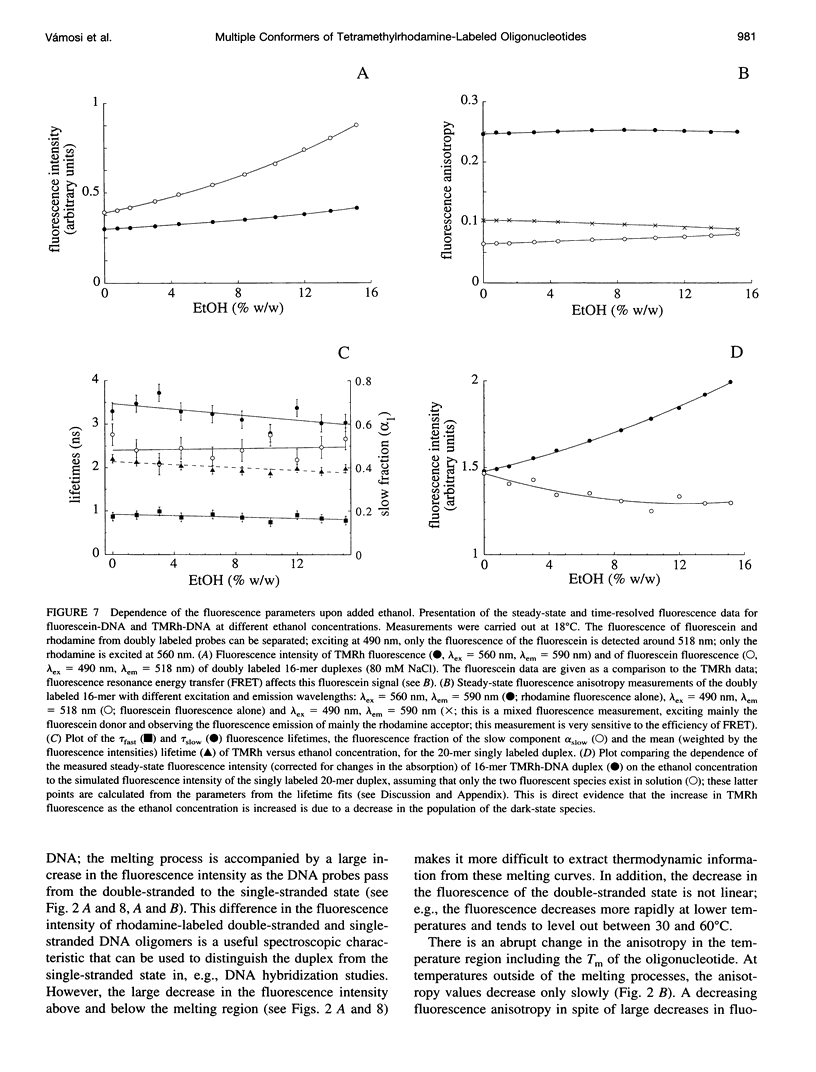
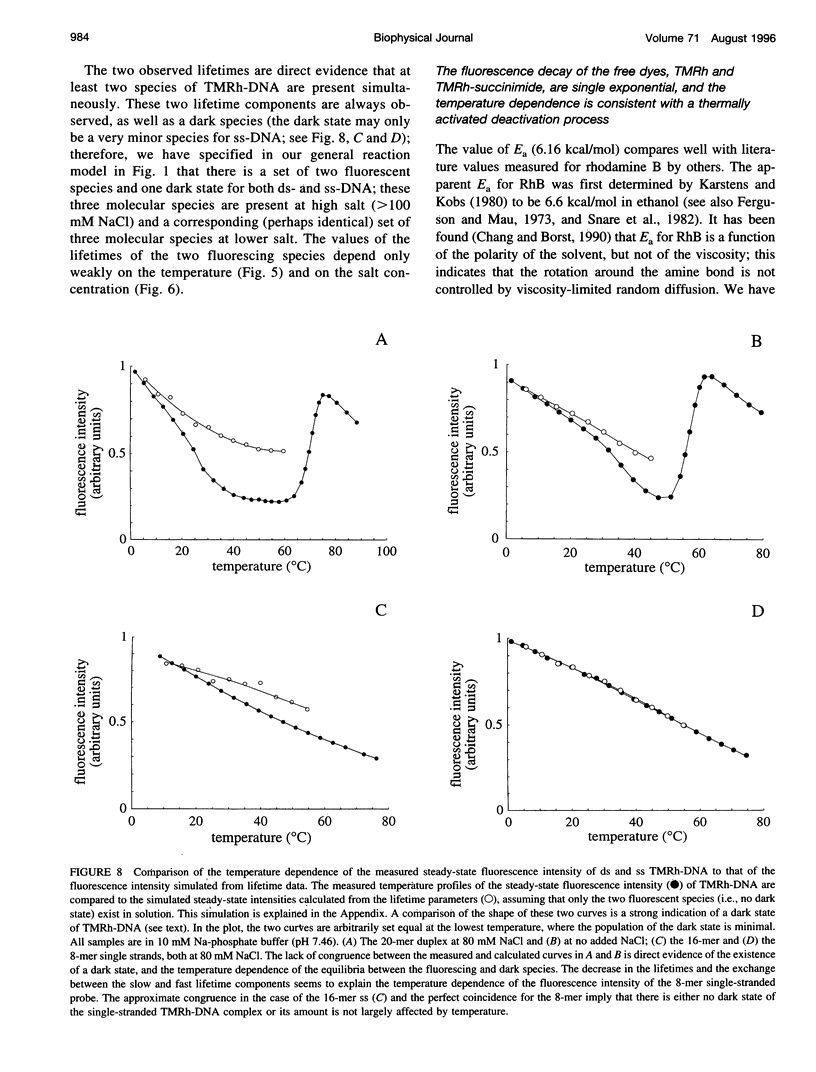
Fluorescence steady-state and lifetime experiments have been carried out on duplex and single-stranded DNA molecules labeled at the 5' ends with 5-carboxytetramethylrhodamine (TMRh). The temperature and ionic strength of the solutions were varied over large ranges. The results reveal at least three well-defined states of the TMRh-DNA molecules for the single-stranded as well as for the double-stranded DNA molecules. Two states are fluorescent, with lifetimes in the range of 0.5-1 ns and 2.5-3 ns. A third state of TMRh-DNA does not fluoresce (a dark species of TMRh-DNA). The distribution of the TMRh-DNA molecules among these three states is strongly temperature and ionic strength dependent. Estimates are made of some reaction parameters of the multistate model. The results are discussed in terms of the photophysics of TMRh, and consequences of the multiple conformers of TMRh-DNA for studies involving fluorescence studies with TMRh-labeled DNA are considered.
Full text
PDF














Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arndt-Jovin D. J., Jovin T. M. Multivariate chromosome analysis and complete karyotyping using dual labeling and fluorescence digital imaging microscopy. Cytometry. 1990;11(1):80–93. doi: 10.1002/cyto.990110110. [DOI] [PubMed] [Google Scholar]
- Boltz R. C., Fischer P. A., Wicker L. S., Peterson L. B. Single UV excitation of Hoechst 33342 and ethidium bromide for simultaneous cell cycle analysis and viability determinations on in vitro cultures of murine B lymphocytes. Cytometry. 1994 Jan 1;15(1):28–34. doi: 10.1002/cyto.990150106. [DOI] [PubMed] [Google Scholar]
- Borgwardt G., Mau H. Prophylaxe des postoperativen Frühileus bei Kindern durch intestianle Schienung. Zentralbl Chir. 1973 Nov 9;98(45):1614–1617. [PubMed] [Google Scholar]
- Breslauer K. J., Frank R., Blöcker H., Marky L. A. Predicting DNA duplex stability from the base sequence. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresloff J. L., Crothers D. M. DNA-ethidium reaction kinetics: demonstration of direct ligand transfer between DNA binding sites. J Mol Biol. 1975 Jun 15;95(1):103–123. doi: 10.1016/0022-2836(75)90339-3. [DOI] [PubMed] [Google Scholar]
- Cardullo R. A., Agrawal S., Flores C., Zamecnik P. C., Wolf D. E. Detection of nucleic acid hybridization by nonradiative fluorescence resonance energy transfer. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8790–8794. doi: 10.1073/pnas.85.23.8790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg R. M. Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol. 1992;211:353–388. doi: 10.1016/0076-6879(92)11020-j. [DOI] [PubMed] [Google Scholar]
- Clegg R. M. Fluorescence resonance energy transfer. Curr Opin Biotechnol. 1995 Feb;6(1):103–110. doi: 10.1016/0958-1669(95)80016-6. [DOI] [PubMed] [Google Scholar]
- Clegg R. M., Murchie A. I., Lilley D. M. The four-way DNA junction: a fluorescence resonance energy transfer study. Braz J Med Biol Res. 1993 Apr;26(4):405–416. [PubMed] [Google Scholar]
- Clegg R. M., Murchie A. I., Lilley D. M. The solution structure of the four-way DNA junction at low-salt conditions: a fluorescence resonance energy transfer analysis. Biophys J. 1994 Jan;66(1):99–109. doi: 10.1016/S0006-3495(94)80765-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg R. M., Murchie A. I., Zechel A., Carlberg C., Diekmann S., Lilley D. M. Fluorescence resonance energy transfer analysis of the structure of the four-way DNA junction. Biochemistry. 1992 May 26;31(20):4846–4856. doi: 10.1021/bi00135a016. [DOI] [PubMed] [Google Scholar]
- Clegg R. M., Murchie A. I., Zechel A., Lilley D. M. Observing the helical geometry of double-stranded DNA in solution by fluorescence resonance energy transfer. Proc Natl Acad Sci U S A. 1993 Apr 1;90(7):2994–2998. doi: 10.1073/pnas.90.7.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly B. A. The synthesis of oligonucleotides containing a primary amino group at the 5'-terminus. Nucleic Acids Res. 1987 Apr 10;15(7):3131–3139. doi: 10.1093/nar/15.7.3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J. P., Hagerman P. J. Analysis of fluorescence energy transfer in duplex and branched DNA molecules. Biochemistry. 1990 Oct 2;29(39):9261–9268. doi: 10.1021/bi00491a022. [DOI] [PubMed] [Google Scholar]
- Dougherty G., Pigram W. J. Spectroscopic analysis of drug-nucleic acid interactions. CRC Crit Rev Biochem. 1982 Feb;12(2):103–132. doi: 10.3109/10409238209108704. [DOI] [PubMed] [Google Scholar]
- Eis P. S., Millar D. P. Conformational distributions of a four-way DNA junction revealed by time-resolved fluorescence resonance energy transfer. Biochemistry. 1993 Dec 21;32(50):13852–13860. doi: 10.1021/bi00213a014. [DOI] [PubMed] [Google Scholar]
- Gohlke C., Murchie A. I., Lilley D. M., Clegg R. M. Kinking of DNA and RNA helices by bulged nucleotides observed by fluorescence resonance energy transfer. Proc Natl Acad Sci U S A. 1994 Nov 22;91(24):11660–11664. doi: 10.1073/pnas.91.24.11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas E., Katchalski-Katzir E., Steinberg I. Z. Effect of the orientation of donor and acceptor on the probability of energy transfer involving electronic transitions of mixed polarization. Biochemistry. 1978 Nov 14;17(23):5064–5070. doi: 10.1021/bi00616a032. [DOI] [PubMed] [Google Scholar]
- Hirons G. T., Fawcett J. J., Crissman H. A. TOTO and YOYO: new very bright fluorochromes for DNA content analyses by flow cytometry. Cytometry. 1994 Feb 1;15(2):129–140. doi: 10.1002/cyto.990150206. [DOI] [PubMed] [Google Scholar]
- Hochstrasser R. A., Chen S. M., Millar D. P. Distance distribution in a dye-linked oligonucleotide determined by time-resolved fluorescence energy transfer. Biophys Chem. 1992 Dec;45(2):133–141. doi: 10.1016/0301-4622(92)87005-4. [DOI] [PubMed] [Google Scholar]
- Jares-Erijman E. A., Jovin T. M. Determination of DNA helical handedness by fluorescence resonance energy transfer. J Mol Biol. 1996 Apr 5;257(3):597–617. doi: 10.1006/jmbi.1996.0188. [DOI] [PubMed] [Google Scholar]
- LePecq J. B., Paoletti C. A fluorescent complex between ethidium bromide and nucleic acids. Physical-chemical characterization. J Mol Biol. 1967 Jul 14;27(1):87–106. doi: 10.1016/0022-2836(67)90353-1. [DOI] [PubMed] [Google Scholar]
- Lilley D. M., Clegg R. M. The structure of branched DNA species. Q Rev Biophys. 1993 May;26(2):131–175. doi: 10.1017/s0033583500004054. [DOI] [PubMed] [Google Scholar]
- Loontiens F. G., McLaughlin L. W., Diekmann S., Clegg R. M. Binding of Hoechst 33258 and 4',6'-diamidino-2-phenylindole to self-complementary decadeoxynucleotides with modified exocyclic base substituents. Biochemistry. 1991 Jan 8;30(1):182–189. doi: 10.1021/bi00215a027. [DOI] [PubMed] [Google Scholar]
- Loontiens F. G., Regenfuss P., Zechel A., Dumortier L., Clegg R. M. Binding characteristics of Hoechst 33258 with calf thymus DNA, poly[d(A-T)], and d(CCGGAATTCCGG): multiple stoichiometries and determination of tight binding with a wide spectrum of site affinities. Biochemistry. 1990 Sep 25;29(38):9029–9039. doi: 10.1021/bi00490a021. [DOI] [PubMed] [Google Scholar]
- Macgregor R. B., Jr, Clegg R. M., Jovin T. M. Pressure-jump study of the kinetics of ethidium bromide binding to DNA. Biochemistry. 1985 Sep 24;24(20):5503–5510. doi: 10.1021/bi00341a034. [DOI] [PubMed] [Google Scholar]
- Macgregor R. B., Jr, Clegg R. M., Jovin T. M. Viscosity dependence of ethidium-DNA intercalation kinetics. Biochemistry. 1987 Jun 30;26(13):4008–4016. doi: 10.1021/bi00387a040. [DOI] [PubMed] [Google Scholar]
- Mergny J. L., Boutorine A. S., Garestier T., Belloc F., Rougée M., Bulychev N. V., Koshkin A. A., Bourson J., Lebedev A. V., Valeur B. Fluorescence energy transfer as a probe for nucleic acid structures and sequences. Nucleic Acids Res. 1994 Mar 25;22(6):920–928. doi: 10.1093/nar/22.6.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison L. E., Halder T. C., Stols L. M. Solution-phase detection of polynucleotides using interacting fluorescent labels and competitive hybridization. Anal Biochem. 1989 Dec;183(2):231–244. doi: 10.1016/0003-2697(89)90473-9. [DOI] [PubMed] [Google Scholar]
- Murchie A. I., Clegg R. M., von Kitzing E., Duckett D. R., Diekmann S., Lilley D. M. Fluorescence energy transfer shows that the four-way DNA junction is a right-handed cross of antiparallel molecules. Nature. 1989 Oct 26;341(6244):763–766. doi: 10.1038/341763a0. [DOI] [PubMed] [Google Scholar]
- Record M. T., Jr, Anderson C. F., Lohman T. M. Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: the roles of ion association or release, screening, and ion effects on water activity. Q Rev Biophys. 1978 May;11(2):103–178. doi: 10.1017/s003358350000202x. [DOI] [PubMed] [Google Scholar]
- Ryan D. P., Crothers D. M. Relaxation kinetics of DNA-ligand binding including direct transfer. Biopolymers. 1984 Mar;23(3):537–562. doi: 10.1002/bip.360230309. [DOI] [PubMed] [Google Scholar]
- Selvin P. R. Fluorescence resonance energy transfer. Methods Enzymol. 1995;246:300–334. doi: 10.1016/0076-6879(95)46015-2. [DOI] [PubMed] [Google Scholar]
- Selvin P. R., Hearst J. E. Luminescence energy transfer using a terbium chelate: improvements on fluorescence energy transfer. Proc Natl Acad Sci U S A. 1994 Oct 11;91(21):10024–10028. doi: 10.1073/pnas.91.21.10024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sixou S., Szoka F. C., Jr, Green G. A., Giusti B., Zon G., Chin D. J. Intracellular oligonucleotide hybridization detected by fluorescence resonance energy transfer (FRET). Nucleic Acids Res. 1994 Feb 25;22(4):662–668. doi: 10.1093/nar/22.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoll V. S., Blanchard J. S. Buffers: principles and practice. Methods Enzymol. 1990;182:24–38. doi: 10.1016/0076-6879(90)82006-n. [DOI] [PubMed] [Google Scholar]
- Tuschl T., Gohlke C., Jovin T. M., Westhof E., Eckstein F. A three-dimensional model for the hammerhead ribozyme based on fluorescence measurements. Science. 1994 Nov 4;266(5186):785–789. doi: 10.1126/science.7973630. [DOI] [PubMed] [Google Scholar]
- Wu P., Brand L. Orientation factor in steady-state and time-resolved resonance energy transfer measurements. Biochemistry. 1992 Sep 1;31(34):7939–7947. doi: 10.1021/bi00149a027. [DOI] [PubMed] [Google Scholar]
- Zimmer C., Wähnert U. Nonintercalating DNA-binding ligands: specificity of the interaction and their use as tools in biophysical, biochemical and biological investigations of the genetic material. Prog Biophys Mol Biol. 1986;47(1):31–112. doi: 10.1016/0079-6107(86)90005-2. [DOI] [PubMed] [Google Scholar]



