Abstract
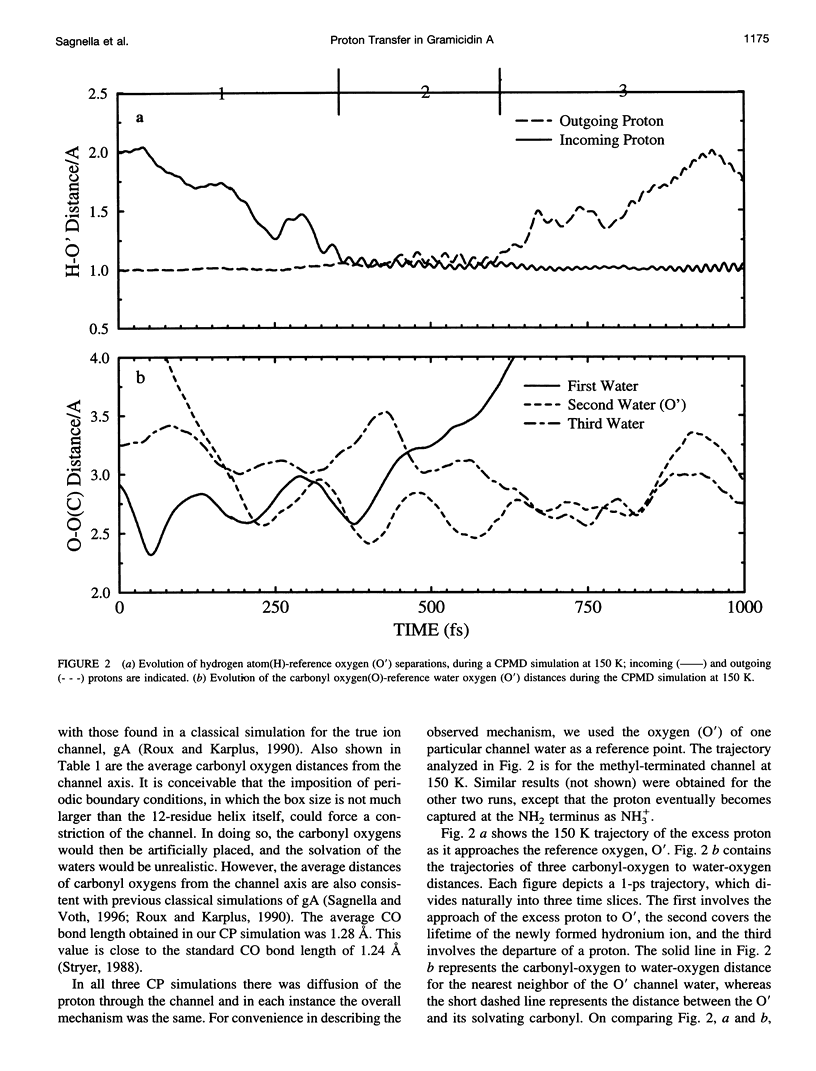
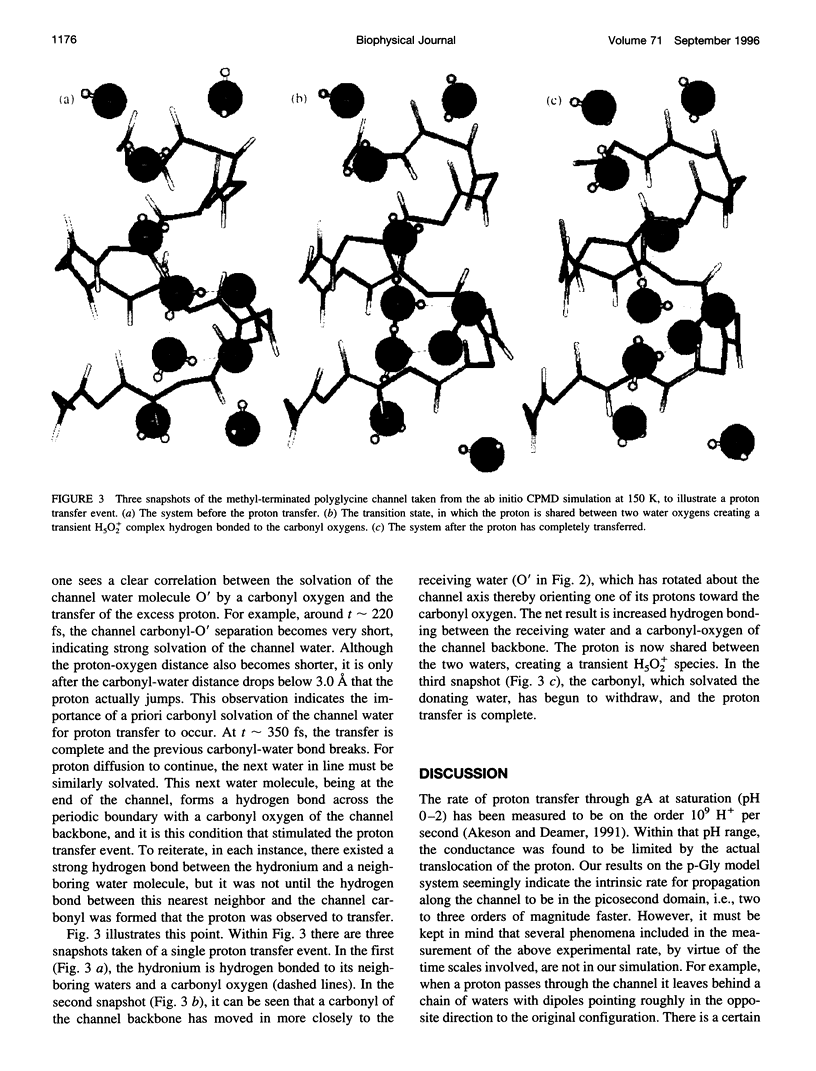
Proton transfer in biological systems is thought to often proceed through hydrogen-bonded chains of water molecules. The ion channel, gramicidin A (gA), houses within its helical structure just such a chain. Using the density functional theory based ab initio molecular dynamics Car-Parrinello method, the structure and dynamics of proton diffusion through a polyglycine analog of the gA ion channel has been investigated. In the channel, a proton, which is initially present as hydronium (H3O+), rapidly forms a strong hydrogen bond with a nearest neighbor water, yielding a transient H5O2+ complex. As in bulk water, strong hydrogen bonding of this complex to a second neighbor solvation shell is required for proton transfer to occur. Within gA, this second neighbor shell included not only a channel water molecule but also a carbonyl of the channel backbone. The present calculations suggest a transport mechanism in which a priori carbonyl solvation is a requirement for proton transfer.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akeson M., Deamer D. W. Proton conductance by the gramicidin water wire. Model for proton conductance in the F1F0 ATPases? Biophys J. 1991 Jul;60(1):101–109. doi: 10.1016/S0006-3495(91)82034-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aqvist J., Warshel A. Energetics of ion permeation through membrane channels. Solvation of Na+ by gramicidin A. Biophys J. 1989 Jul;56(1):171–182. doi: 10.1016/S0006-3495(89)82662-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becke AD. Density-functional exchange-energy approximation with correct asymptotic behavior. Phys Rev A Gen Phys. 1988 Sep 15;38(6):3098–3100. doi: 10.1103/physreva.38.3098. [DOI] [PubMed] [Google Scholar]
- Becker M. D., Koeppe R. E., 2nd, Andersen O. S. Amino acid substitutions and ion channel function. Model-dependent conclusions. Biophys J. 1992 Apr;62(1):25–27. doi: 10.1016/S0006-3495(92)81767-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benamar D., Daumas P., Trudelle Y., Calas B., Bennes R., Heitz F. Influence of the nature of the aromatic side-chain on the conductance of the channel of linear gramicidin: study of a series of 9,11,13,15-Tyr(O-protected) derivatives. Eur Biophys J. 1993;22(2):145–150. doi: 10.1007/BF00196918. [DOI] [PubMed] [Google Scholar]
- Cao Y., Váró G., Chang M., Ni B. F., Needleman R., Lanyi J. K. Water is required for proton transfer from aspartate-96 to the bacteriorhodopsin Schiff base. Biochemistry. 1991 Nov 12;30(45):10972–10979. doi: 10.1021/bi00109a023. [DOI] [PubMed] [Google Scholar]
- Car R, Parrinello M. Unified approach for molecular dynamics and density-functional theory. Phys Rev Lett. 1985 Nov 25;55(22):2471–2474. doi: 10.1103/PhysRevLett.55.2471. [DOI] [PubMed] [Google Scholar]
- Chiu S. W., Jakobsson E., Subramaniam S., McCammon J. A. Time-correlation analysis of simulated water motion in flexible and rigid gramicidin channels. Biophys J. 1991 Jul;60(1):273–285. doi: 10.1016/S0006-3495(91)82049-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu S. W., Subramaniam S., Jakobsson E., McCammon J. A. Water and polypeptide conformations in the gramicidin channel. A molecular dynamics study. Biophys J. 1989 Aug;56(2):253–261. doi: 10.1016/S0006-3495(89)82671-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisenhofer J., Michel H. Nobel lecture. The photosynthetic reaction centre from the purple bacterium Rhodopseudomonas viridis. EMBO J. 1989 Aug;8(8):2149–2170. doi: 10.1002/j.1460-2075.1989.tb08338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein A., Andersen O. S. The gramicidin A channel: a review of its permeability characteristics with special reference to the single-file aspect of transport. J Membr Biol. 1981 Apr 30;59(3):155–171. doi: 10.1007/BF01875422. [DOI] [PubMed] [Google Scholar]
- Henderson R., Baldwin J. M., Ceska T. A., Zemlin F., Beckmann E., Downing K. H. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol. 1990 Jun 20;213(4):899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- Hu W., Cross T. A. Tryptophan hydrogen bonding and electric dipole moments: functional roles in the gramicidin channel and implications for membrane proteins. Biochemistry. 1995 Oct 31;34(43):14147–14155. doi: 10.1021/bi00043a020. [DOI] [PubMed] [Google Scholar]
- Laasonen K, Pasquarello A, Car R, Lee C, Vanderbilt D. Car-Parrinello molecular dynamics with Vanderbilt ultrasoft pseudopotentials. Phys Rev B Condens Matter. 1993 Apr 15;47(16):10142–10153. doi: 10.1103/physrevb.47.10142. [DOI] [PubMed] [Google Scholar]
- Lee W. K., Jordan P. C. Molecular dynamics simulation of cation motion in water-filled gramicidinlike pores. Biophys J. 1984 Dec;46(6):805–819. doi: 10.1016/S0006-3495(84)84079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt D. G., Elias S. R., Hautman J. M. Number of water molecules coupled to the transport of sodium, potassium and hydrogen ions via gramicidin, nonactin or valinomycin. Biochim Biophys Acta. 1978 Sep 22;512(2):436–451. doi: 10.1016/0005-2736(78)90266-3. [DOI] [PubMed] [Google Scholar]
- Nagle J. F., Morowitz H. J. Molecular mechanisms for proton transport in membranes. Proc Natl Acad Sci U S A. 1978 Jan;75(1):298–302. doi: 10.1073/pnas.75.1.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux B., Karplus M. Ion transport in a model gramicidin channel. Structure and thermodynamics. Biophys J. 1991 May;59(5):961–981. doi: 10.1016/S0006-3495(91)82311-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagnella D. E., Voth G. A. Structure and dynamics of hydronium in the ion channel gramicidin A. Biophys J. 1996 May;70(5):2043–2051. doi: 10.1016/S0006-3495(96)79773-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban B. W., Hladky S. B. Ion transport in the simplest single file pore. Biochim Biophys Acta. 1979 Jul 5;554(2):410–429. doi: 10.1016/0005-2736(79)90381-x. [DOI] [PubMed] [Google Scholar]
- Urry D. W. The gramicidin A transmembrane channel: a proposed pi(L,D) helix. Proc Natl Acad Sci U S A. 1971 Mar;68(3):672–676. doi: 10.1073/pnas.68.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Mau N., Bonnet B., Benayad A., Heitz F. The conformation of linear gramicidin is sequence dependent. A monolayer and infrared study. Eur Biophys J. 1994;22(6):447–452. doi: 10.1007/BF00180165. [DOI] [PubMed] [Google Scholar]
- Vanderbilt D. Soft self-consistent pseudopotentials in a generalized eigenvalue formalism. Phys Rev B Condens Matter. 1990 Apr 15;41(11):7892–7895. doi: 10.1103/physrevb.41.7892. [DOI] [PubMed] [Google Scholar]