Abstract
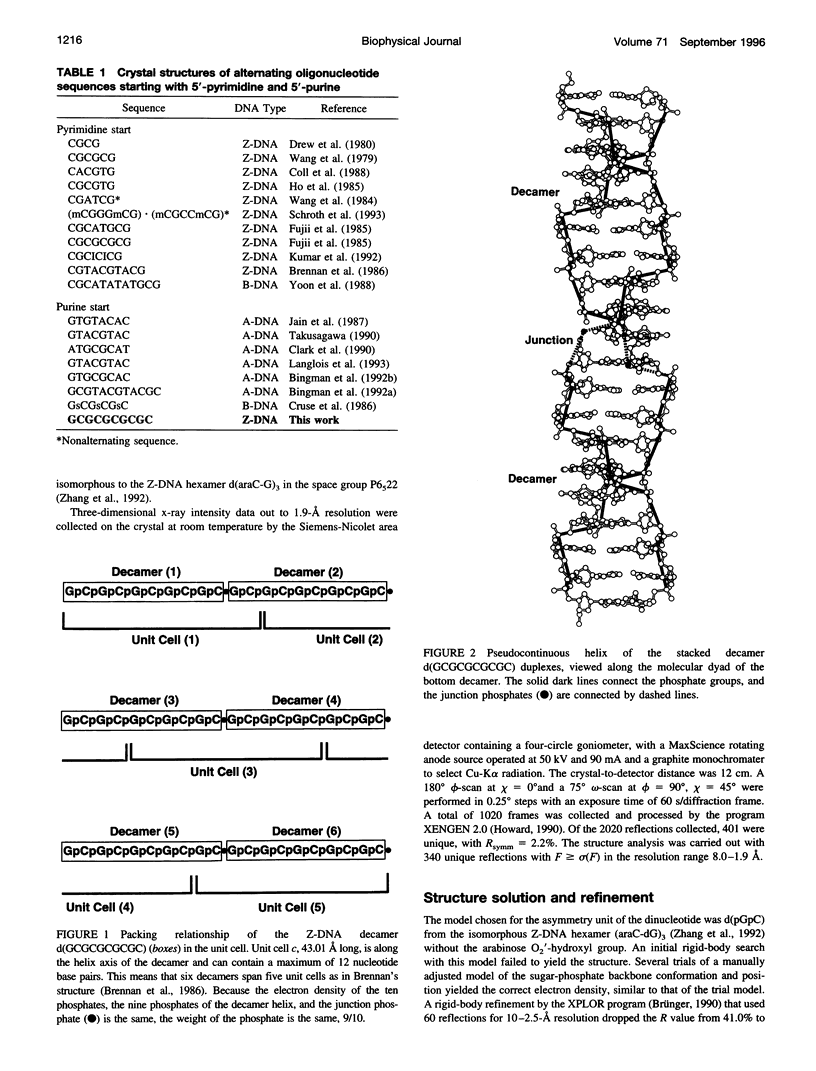
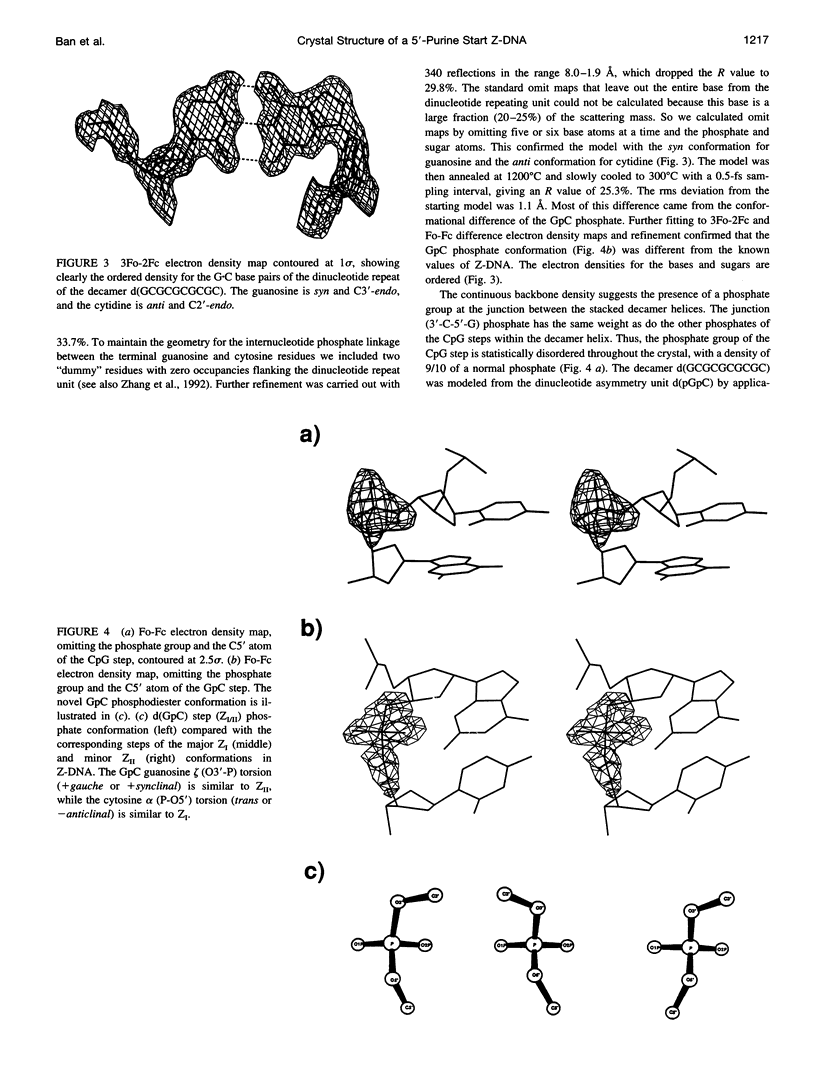
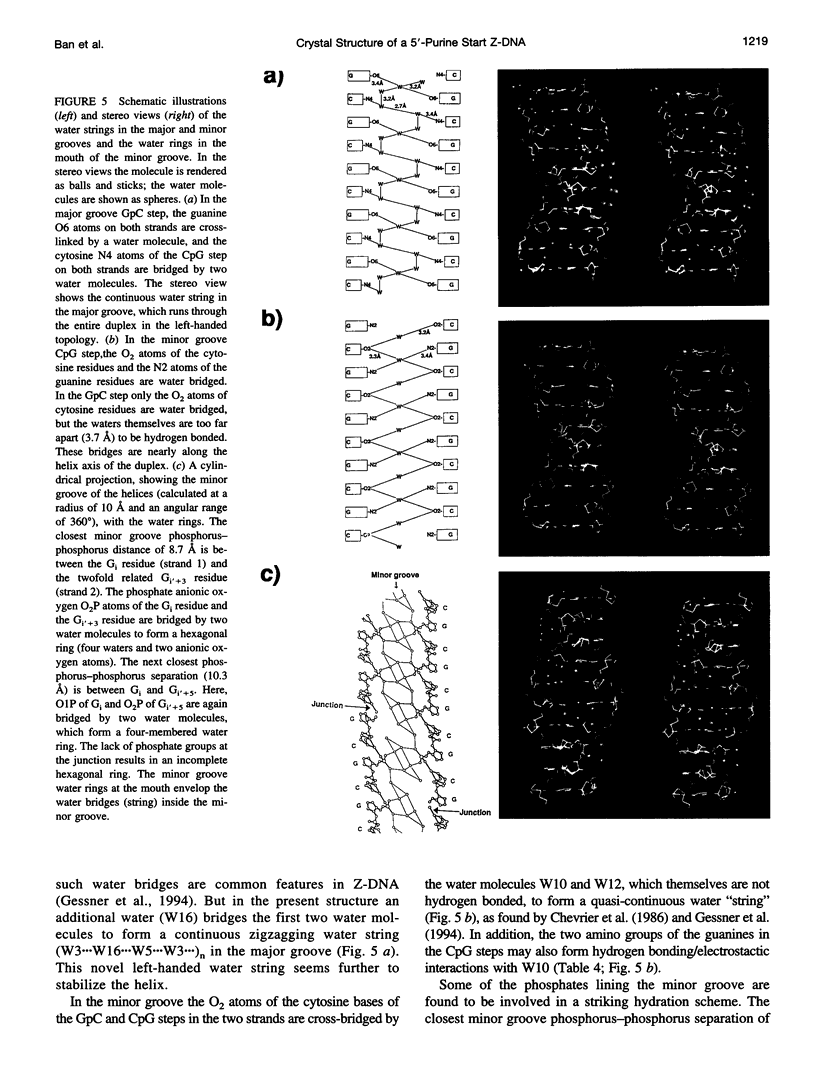
Alternating self-complementary oligonucleotides starting with a 5'-pyrimidine usually form left-handed Z-DNA; however, with a 5'-purine start sequence they form the right-handed A-DNA. Here we report the crystal structure of the decamer d(GCGCGCGCGC) with a 5'-purine start in the Z-DNA form. The decamer crystallizes in the hexagonal space group P6(5)22, unit cell dimensions a = b = 18.08 and c = 43.10 A, with one of the following four dinucleotide diphosphates in the asymmetric unit: d(pGpC)/d(GpCp)/d(pCpG)/d(CpGp). The molecular replacement method, starting with d(pGpC) of the isomorphous Z-DNA hexamer d(araC-dG)3 without the 2'-OH group of arabinose, was used in the structure analysis. The method gave the solution only after the sugar-phosphate conformation of the GpC step was manipulated. The refinement converged to a final R value of 18.6% for 340 unique reflections in the resolution range 8.0-1.9 A. A result of the sequence alternation is the alternation in the nucleotide conformation; guanosine is C3'-endo, syn, and cytidine is C2'-endo, anti. The CpG step phosphodiester conformation is the same as ZI or ZII, whereas that of the GpC step phosphodiester is "intermediate" in the sense that zeta (O3'-P bond) is the same as ZII but alpha (P-O5' bond) is the same as ZI. The duplexes generated from the dinucleotide asymmetric unit are stacked one on top of the other in the crystal to form an infinite pseudocontinuous helix. This renders it a quasi-polymerlike structure that has assumed the Z-DNA conformation further strengthened by the long inner Z-forming stretch d(CG)4. An interesting feature of the structure is the presence of water strings in both the major and the minor grooves. In the minor groove the cytosine carbonyl oxygen atoms of the GpC and CpG steps are cross-bridged by water molecules that are not themselves hydrogen bonded but are enclosed by the water rings in the mouth of the minor groove. In the major groove three independent water molecules form a zigzagging continuous water string that runs throughout the duplex.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Chandrasekaran R., Birdsall D. L., Leslie A. G., Ratliff R. L. Left-handed DNA helices. Nature. 1980 Feb 21;283(5749):743–745. doi: 10.1038/283743a0. [DOI] [PubMed] [Google Scholar]
- Berman H. M., Olson W. K., Beveridge D. L., Westbrook J., Gelbin A., Demeny T., Hsieh S. H., Srinivasan A. R., Schneider B. The nucleic acid database. A comprehensive relational database of three-dimensional structures of nucleic acids. Biophys J. 1992 Sep;63(3):751–759. doi: 10.1016/S0006-3495(92)81649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingman C., Jain S., Zon G., Sundaralingam M. Crystal and molecular structure of the alternating dodecamer d(GCGTACGTACGC) in the A-DNA form: comparison with the isomorphous non-alternating dodecamer d(CCGTACGTACGG). Nucleic Acids Res. 1992 Dec 25;20(24):6637–6647. doi: 10.1093/nar/20.24.6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan R. G., Westhof E., Sundaralingam M. Structure of a Z-DNA with two different backbone chain conformations. Stabilization of the decadeoxyoligonucleotide d(CGTACGTACG) by [Co(NH3)6]3+ binding to the guanine. J Biomol Struct Dyn. 1986 Feb;3(4):649–665. doi: 10.1080/07391102.1986.10508453. [DOI] [PubMed] [Google Scholar]
- Chevrier B., Dock A. C., Hartmann B., Leng M., Moras D., Thuong M. T., Westhof E. Solvation of the left-handed hexamer d(5BrC-G-5BrC-G-5 BrC-G) in crystals grown at two temperatures. J Mol Biol. 1986 Apr 20;188(4):707–719. doi: 10.1016/s0022-2836(86)80016-x. [DOI] [PubMed] [Google Scholar]
- Clark G. R., Brown D. G., Sanderson M. R., Chwalinski T., Neidle S., Veal J. M., Jones R. L., Wilson W. D., Zon G., Garman E. Crystal and solution structures of the oligonucleotide d(ATGCGCAT)2: a combined X-ray and NMR study. Nucleic Acids Res. 1990 Sep 25;18(18):5521–5528. doi: 10.1093/nar/18.18.5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll M., Fita I., Lloveras J., Subirana J. A., Bardella F., Huynh-Dinh T., Igolen J. Structure of d(CACGTG), a Z-DNA hexamer containing AT base pairs. Nucleic Acids Res. 1988 Sep 12;16(17):8695–8705. doi: 10.1093/nar/16.17.8695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruse W. B., Salisbury S. A., Brown T., Cosstick R., Eckstein F., Kennard O. Chiral phosphorothioate analogues of B-DNA. The crystal structure of Rp-d[Gp(S)CpGp(S)CpGp(S)C]. J Mol Biol. 1986 Dec 20;192(4):891–905. doi: 10.1016/0022-2836(86)90035-5. [DOI] [PubMed] [Google Scholar]
- Drew H., Takano T., Tanaka S., Itakura K., Dickerson R. E. High-salt d(CpGpCpG), a left-handed Z' DNA double helix. Nature. 1980 Aug 7;286(5773):567–573. doi: 10.1038/286567a0. [DOI] [PubMed] [Google Scholar]
- Fujii S., Wang A. H., Quigley G. J., Westerink H., Van der Marel G., Van Boom J. H., Rich A. The octamers d(CGCGCGCG) and d(CGCATGCG) both crystallize as Z-DNA in the same hexagonal lattice. Biopolymers. 1985 Jan;24(1):243–250. doi: 10.1002/bip.360240118. [DOI] [PubMed] [Google Scholar]
- Gessner R. V., Quigley G. J., Egli M. Comparative studies of high resolution Z-DNA crystal structures. Part 1: Common hydration patterns of alternating dC-dG. J Mol Biol. 1994 Mar 4;236(4):1154–1168. doi: 10.1016/0022-2836(94)90018-3. [DOI] [PubMed] [Google Scholar]
- Ho P. S., Frederick C. A., Quigley G. J., van der Marel G. A., van Boom J. H., Wang A. H., Rich A. G.T wobble base-pairing in Z-DNA at 1.0 A atomic resolution: the crystal structure of d(CGCGTG). EMBO J. 1985 Dec 16;4(13A):3617–3623. doi: 10.1002/j.1460-2075.1985.tb04125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S., Zon G., Sundaralingam M. The potentially Z-DNA-forming sequence d(GTGTACAC) crystallizes as A-DNA. J Mol Biol. 1987 Sep 5;197(1):141–145. doi: 10.1016/0022-2836(87)90616-4. [DOI] [PubMed] [Google Scholar]
- Kumar V. D., Harrison R. W., Andrews L. C., Weber I. T. Crystal structure at 1.5-A resolution of d(CGCICICG), an octanucleotide containing inosine, and its comparison with d(CGCG) and d(CGCGCG) structures. Biochemistry. 1992 Feb 11;31(5):1541–1550. doi: 10.1021/bi00120a035. [DOI] [PubMed] [Google Scholar]
- Langlois d'Estaintot B., Dautant A., Courseille C., Precigoux G. Orthorhombic crystal structure of the A-DNA octamer d(GTACGTAC). Comparison with the tetragonal structure. Eur J Biochem. 1993 Apr 15;213(2):673–682. doi: 10.1111/j.1432-1033.1993.tb17807.x. [DOI] [PubMed] [Google Scholar]
- Mooers B. H., Schroth G. P., Baxter W. W., Ho P. S. Alternating and non-alternating dG-dC hexanucleotides crystallize as canonical A-DNA. J Mol Biol. 1995 Jun 16;249(4):772–784. doi: 10.1006/jmbi.1995.0336. [DOI] [PubMed] [Google Scholar]
- Quadrifoglio F., Manzini G., Yathindra N. Short oligodeoxynucleotides with d(G-C)n sequence do not assume left-handed conformation in high salt conditions. J Mol Biol. 1984 May 25;175(3):419–423. doi: 10.1016/0022-2836(84)90358-9. [DOI] [PubMed] [Google Scholar]
- Takusagawa F. The crystal structure of d(GTACGTAC) at 2.25 A resolution: are the A-DNA's always unwound approximately 10 degrees at the C-G steps? J Biomol Struct Dyn. 1990 Feb;7(4):795–809. doi: 10.1080/07391102.1990.10508524. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Hakoshima T., van der Marel G., van Boom J. H., Rich A. AT base pairs are less stable than GC base pairs in Z-DNA: the crystal structure of d(m5CGTAm5CG). Cell. 1984 May;37(1):321–331. doi: 10.1016/0092-8674(84)90328-3. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Wang A. J., Quigley G. J., Kolpak F. J., van der Marel G., van Boom J. H., Rich A. Left-handed double helical DNA: variations in the backbone conformation. Science. 1981 Jan 9;211(4478):171–176. doi: 10.1126/science.7444458. [DOI] [PubMed] [Google Scholar]
- Yoon C., Privé G. G., Goodsell D. S., Dickerson R. E. Structure of an alternating-B DNA helix and its relationship to A-tract DNA. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6332–6336. doi: 10.1073/pnas.85.17.6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., van der Marel G. A., van Boom J. H., Wang A. H. Conformational perturbation of the anticancer nucleotide arabinosylcytosine on Z-DNA: molecular structure of (araC-dG)3 at 1.3 A resolution. Biopolymers. 1992 Nov;32(11):1559–1569. doi: 10.1002/bip.360321113. [DOI] [PubMed] [Google Scholar]



