Abstract
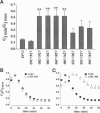
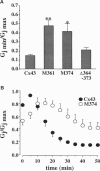
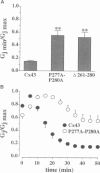
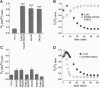
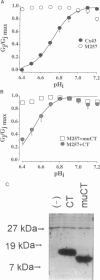
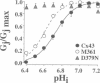
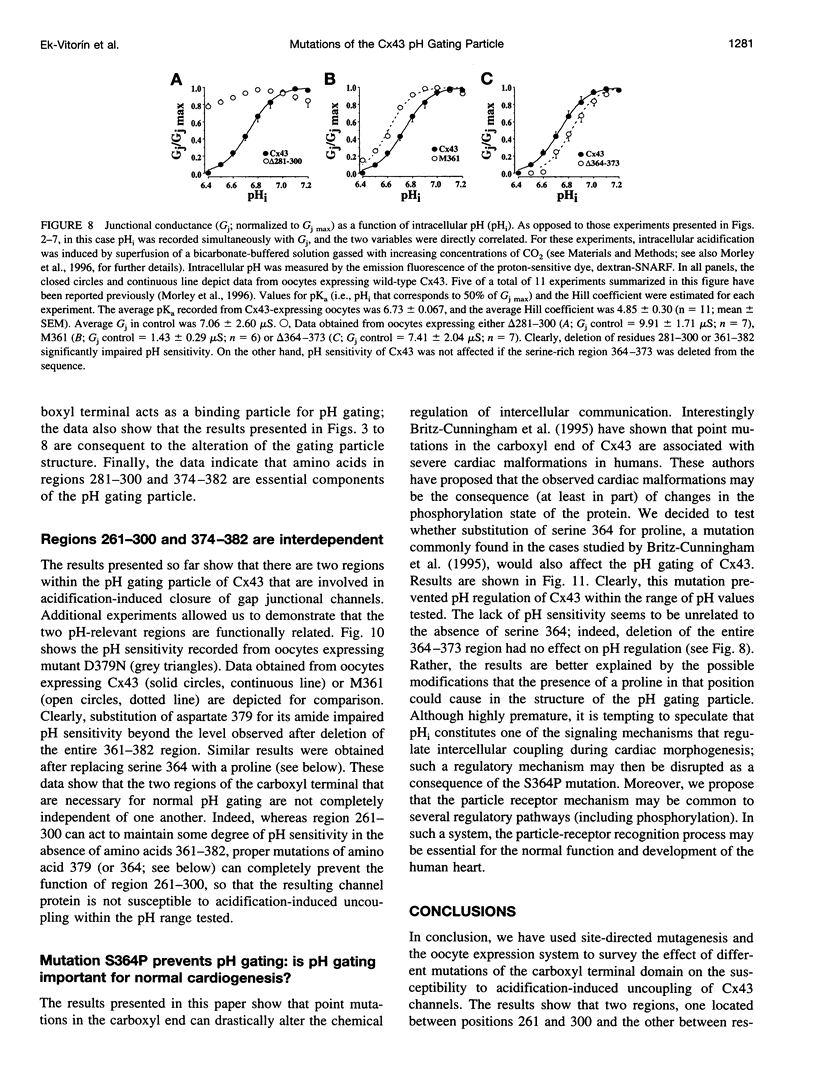
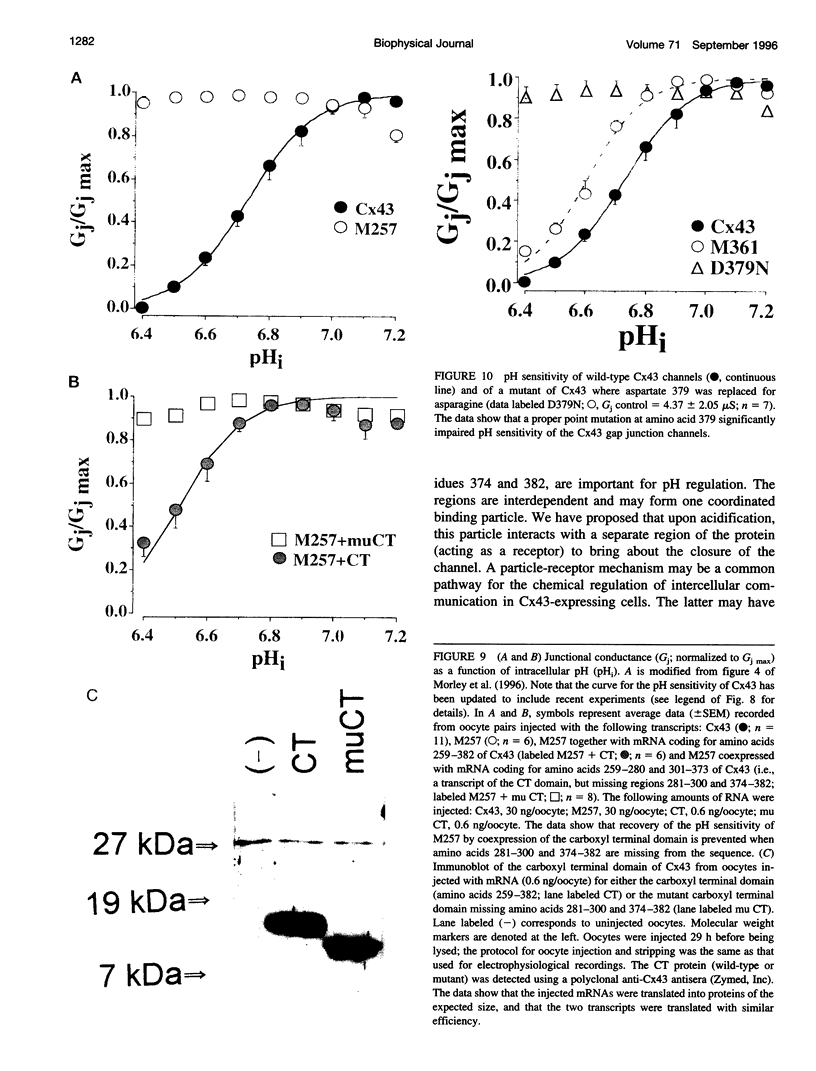
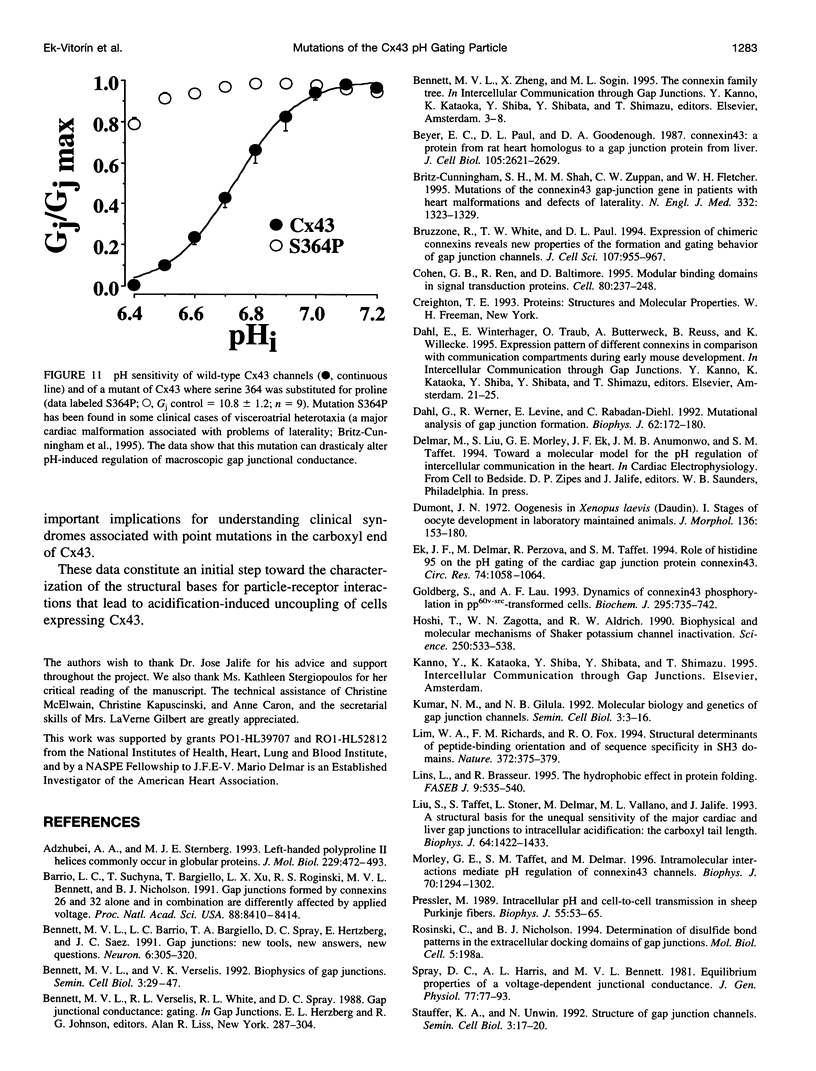
Gap junction channels allow for the passage of ions and small molecules between neighboring cells. These channels are formed by multimers of an integral membrane protein named connexin. In the heart and other tissues, the most abundant connexin is a 43-kDa, 382-amino acid protein termed connexin43 (Cx43). A characteristic property of connexin channels is that they close upon acidification of the intracellular space. Previous studies have shown that truncation of the carboxyl terminal of Cx43 impairs pH sensitivity. In the present study, we have used a combination of optical, electrophysiological, and molecular biological techniques and the oocyte expression system to further localize the regions of the carboxyl terminal that are involved in pH regulation of Cx43 channels. Our results show that regions 261-300 and 374-382 are essential components of a pH-dependent "gating particle," which is responsible for acidification-induced uncoupling of Cx43-expressing cells. Regions 261-300 and 374-382 seem to be interdependent. The function of region 261-300 may be related to the presence of a poly-proline repeat between amino acids 274 and 285. Furthermore, site-directed mutagenesis studies show that the function of region 374-382 is not directly related to its net balance of charges, although mutation of only one amino acid (aspartate 379) for asparagine impairs pH sensitivity to the same extent as truncation of the carboxyl terminal domain (from amino acid 257). The mutation in which serine 364 is substituted for proline, which has been associated with some cases of cardiac congenital malformations in humans, also disrupts the pH gating of Cx43, although deletion of amino acids 364-373 has no effect on acidification-induced uncoupling. These results provide new insight into the molecular mechanisms responsible for acidification-induced uncoupling of gap junction channels in the heart and in other Cx43-expressing structures.
Full text
PDF

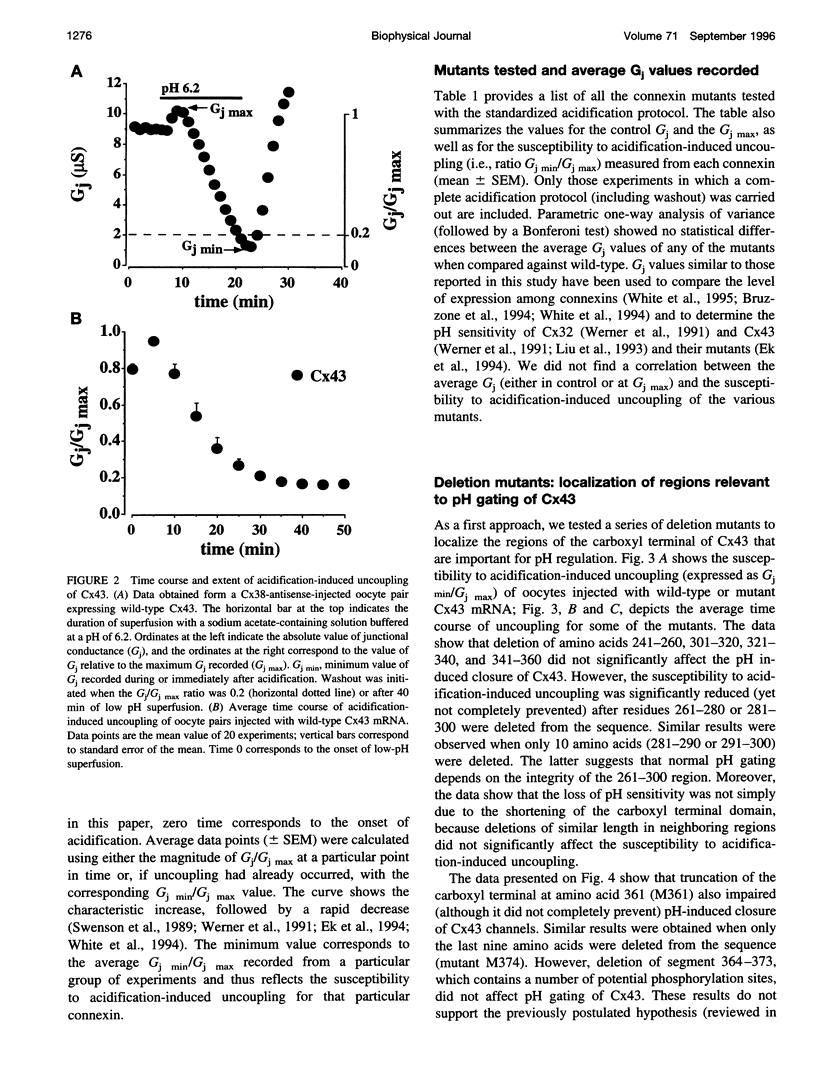
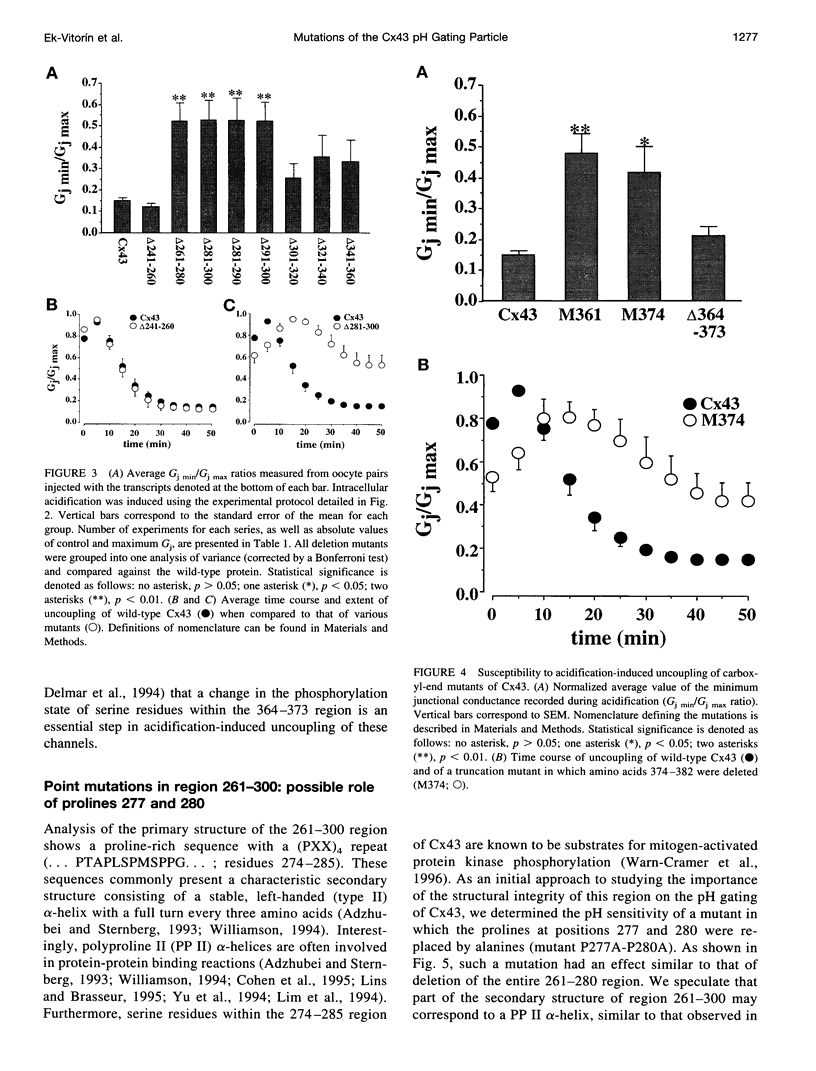
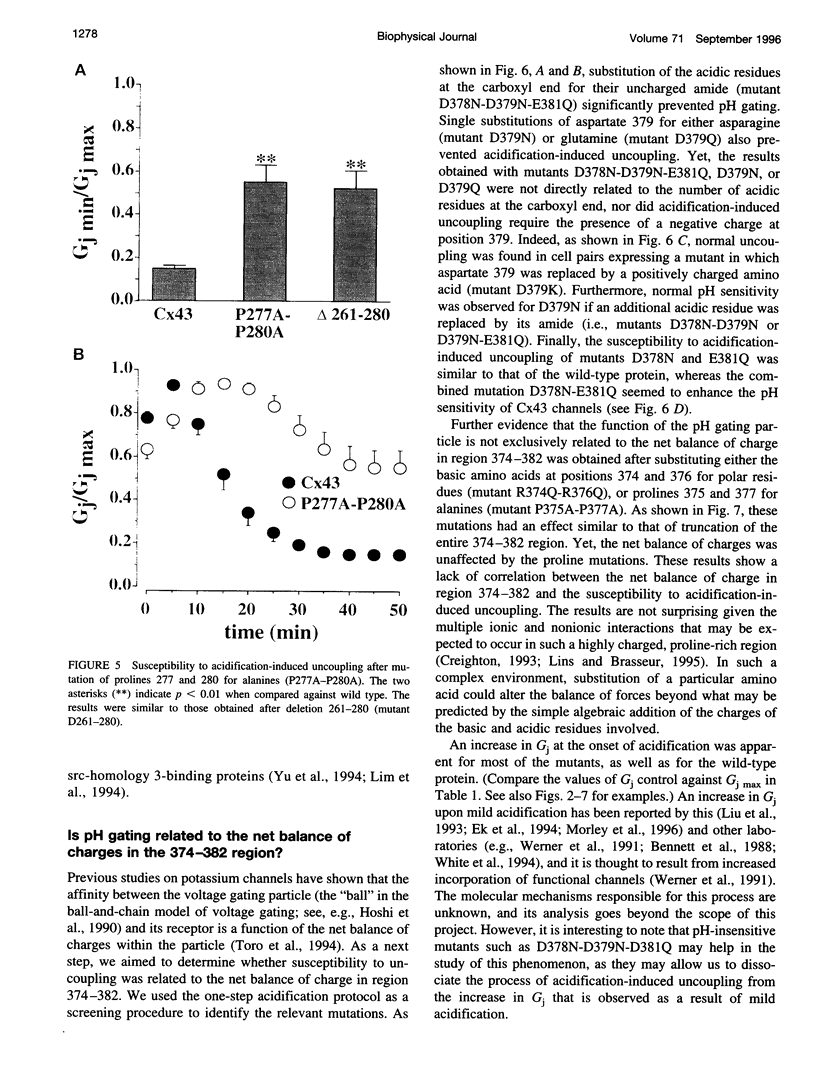
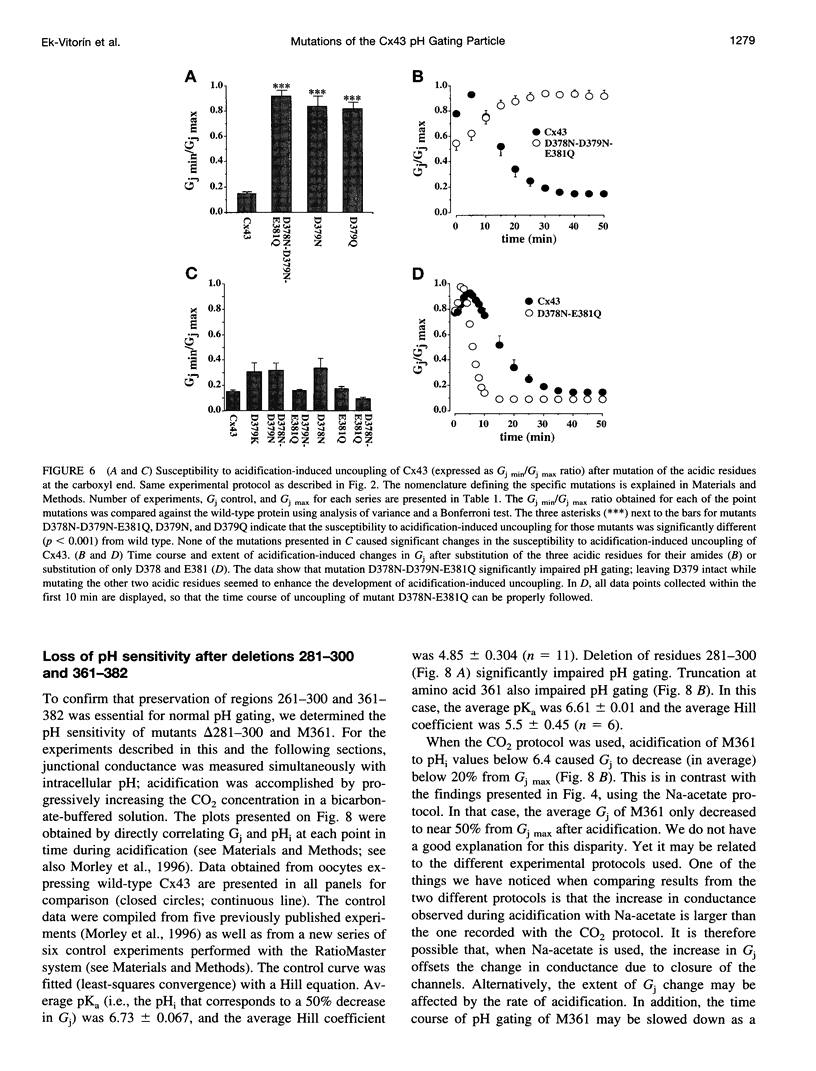
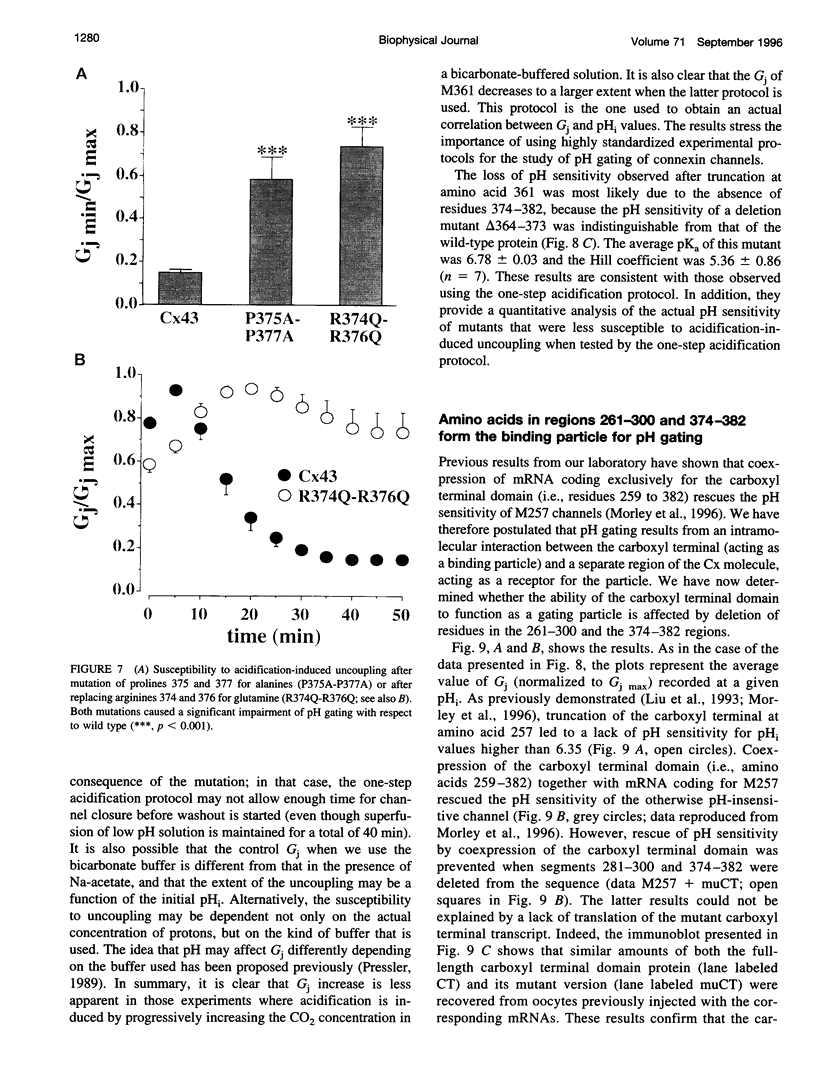

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adzhubei A. A., Sternberg M. J. Left-handed polyproline II helices commonly occur in globular proteins. J Mol Biol. 1993 Jan 20;229(2):472–493. doi: 10.1006/jmbi.1993.1047. [DOI] [PubMed] [Google Scholar]
- Barrio L. C., Suchyna T., Bargiello T., Xu L. X., Roginski R. S., Bennett M. V., Nicholson B. J. Gap junctions formed by connexins 26 and 32 alone and in combination are differently affected by applied voltage. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8410–8414. doi: 10.1073/pnas.88.19.8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M. V., Barrio L. C., Bargiello T. A., Spray D. C., Hertzberg E., Sáez J. C. Gap junctions: new tools, new answers, new questions. Neuron. 1991 Mar;6(3):305–320. doi: 10.1016/0896-6273(91)90241-q. [DOI] [PubMed] [Google Scholar]
- Bennett M. V., Verselis V. K. Biophysics of gap junctions. Semin Cell Biol. 1992 Feb;3(1):29–47. doi: 10.1016/s1043-4682(10)80006-6. [DOI] [PubMed] [Google Scholar]
- Beyer E. C., Paul D. L., Goodenough D. A. Connexin43: a protein from rat heart homologous to a gap junction protein from liver. J Cell Biol. 1987 Dec;105(6 Pt 1):2621–2629. doi: 10.1083/jcb.105.6.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britz-Cunningham S. H., Shah M. M., Zuppan C. W., Fletcher W. H. Mutations of the Connexin43 gap-junction gene in patients with heart malformations and defects of laterality. N Engl J Med. 1995 May 18;332(20):1323–1329. doi: 10.1056/NEJM199505183322002. [DOI] [PubMed] [Google Scholar]
- Bruzzone R., White T. W., Paul D. L. Expression of chimeric connexins reveals new properties of the formation and gating behavior of gap junction channels. J Cell Sci. 1994 Apr;107(Pt 4):955–967. doi: 10.1242/jcs.107.4.955. [DOI] [PubMed] [Google Scholar]
- Cohen G. B., Ren R., Baltimore D. Modular binding domains in signal transduction proteins. Cell. 1995 Jan 27;80(2):237–248. doi: 10.1016/0092-8674(95)90406-9. [DOI] [PubMed] [Google Scholar]
- Dahl G., Werner R., Levine E., Rabadan-Diehl C. Mutational analysis of gap junction formation. Biophys J. 1992 Apr;62(1):172–182. doi: 10.1016/S0006-3495(92)81803-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont J. N. Oogenesis in Xenopus laevis (Daudin). I. Stages of oocyte development in laboratory maintained animals. J Morphol. 1972 Feb;136(2):153–179. doi: 10.1002/jmor.1051360203. [DOI] [PubMed] [Google Scholar]
- Ek J. F., Delmar M., Perzova R., Taffet S. M. Role of histidine 95 on pH gating of the cardiac gap junction protein connexin43. Circ Res. 1994 Jun;74(6):1058–1064. doi: 10.1161/01.res.74.6.1058. [DOI] [PubMed] [Google Scholar]
- Goldberg G. S., Lau A. F. Dynamics of connexin43 phosphorylation in pp60v-src-transformed cells. Biochem J. 1993 Nov 1;295(Pt 3):735–742. doi: 10.1042/bj2950735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshi T., Zagotta W. N., Aldrich R. W. Biophysical and molecular mechanisms of Shaker potassium channel inactivation. Science. 1990 Oct 26;250(4980):533–538. doi: 10.1126/science.2122519. [DOI] [PubMed] [Google Scholar]
- Kumar N. M., Gilula N. B. Molecular biology and genetics of gap junction channels. Semin Cell Biol. 1992 Feb;3(1):3–16. doi: 10.1016/s1043-4682(10)80003-0. [DOI] [PubMed] [Google Scholar]
- Lim W. A., Richards F. M., Fox R. O. Structural determinants of peptide-binding orientation and of sequence specificity in SH3 domains. Nature. 1994 Nov 24;372(6504):375–379. doi: 10.1038/372375a0. [DOI] [PubMed] [Google Scholar]
- Lins L., Brasseur R. The hydrophobic effect in protein folding. FASEB J. 1995 Apr;9(7):535–540. doi: 10.1096/fasebj.9.7.7737462. [DOI] [PubMed] [Google Scholar]
- Liu S., Taffet S., Stoner L., Delmar M., Vallano M. L., Jalife J. A structural basis for the unequal sensitivity of the major cardiac and liver gap junctions to intracellular acidification: the carboxyl tail length. Biophys J. 1993 May;64(5):1422–1433. doi: 10.1016/S0006-3495(93)81508-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley G. E., Taffet S. M., Delmar M. Intramolecular interactions mediate pH regulation of connexin43 channels. Biophys J. 1996 Mar;70(3):1294–1302. doi: 10.1016/S0006-3495(96)79686-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pressler M. L. Intracellular pH and cell-to-cell transmission in sheep Purkinje fibers. Biophys J. 1989 Jan;55(1):53–65. doi: 10.1016/S0006-3495(89)82780-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spray D. C., Harris A. L., Bennett M. V. Equilibrium properties of a voltage-dependent junctional conductance. J Gen Physiol. 1981 Jan;77(1):77–93. doi: 10.1085/jgp.77.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauffer K. A., Unwin N. Structure of gap junction channels. Semin Cell Biol. 1992 Feb;3(1):17–20. doi: 10.1016/s1043-4682(10)80004-2. [DOI] [PubMed] [Google Scholar]
- Suchyna T. M., Xu L. X., Gao F., Fourtner C. R., Nicholson B. J. Identification of a proline residue as a transduction element involved in voltage gating of gap junctions. Nature. 1993 Oct 28;365(6449):847–849. doi: 10.1038/365847a0. [DOI] [PubMed] [Google Scholar]
- Swenson K. I., Jordan J. R., Beyer E. C., Paul D. L. Formation of gap junctions by expression of connexins in Xenopus oocyte pairs. Cell. 1989 Apr 7;57(1):145–155. doi: 10.1016/0092-8674(89)90180-3. [DOI] [PubMed] [Google Scholar]
- Toro L., Ottolia M., Stefani E., Latorre R. Structural determinants in the interaction of Shaker inactivating peptide and a Ca(2+)-activated K+ channel. Biochemistry. 1994 Jun 14;33(23):7220–7228. doi: 10.1021/bi00189a026. [DOI] [PubMed] [Google Scholar]
- Vandeyar M. A., Weiner M. P., Hutton C. J., Batt C. A. A simple and rapid method for the selection of oligodeoxynucleotide-directed mutants. Gene. 1988 May 15;65(1):129–133. doi: 10.1016/0378-1119(88)90425-8. [DOI] [PubMed] [Google Scholar]
- Warn-Cramer B. J., Lampe P. D., Kurata W. E., Kanemitsu M. Y., Loo L. W., Eckhart W., Lau A. F. Characterization of the mitogen-activated protein kinase phosphorylation sites on the connexin-43 gap junction protein. J Biol Chem. 1996 Feb 16;271(7):3779–3786. doi: 10.1074/jbc.271.7.3779. [DOI] [PubMed] [Google Scholar]
- Werner R., Levine E., Rabadan-Diehl C., Dahl G. Gating properties of connexin32 cell-cell channels and their mutants expressed in Xenopus oocytes. Proc Biol Sci. 1991 Jan 22;243(1306):5–11. doi: 10.1098/rspb.1991.0002. [DOI] [PubMed] [Google Scholar]
- White T. W., Bruzzone R., Wolfram S., Paul D. L., Goodenough D. A. Selective interactions among the multiple connexin proteins expressed in the vertebrate lens: the second extracellular domain is a determinant of compatibility between connexins. J Cell Biol. 1994 May;125(4):879–892. doi: 10.1083/jcb.125.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T. W., Paul D. L., Goodenough D. A., Bruzzone R. Functional analysis of selective interactions among rodent connexins. Mol Biol Cell. 1995 Apr;6(4):459–470. doi: 10.1091/mbc.6.4.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson M. P. The structure and function of proline-rich regions in proteins. Biochem J. 1994 Jan 15;297(Pt 2):249–260. doi: 10.1042/bj2970249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Chen J. K., Feng S., Dalgarno D. C., Brauer A. W., Schreiber S. L. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell. 1994 Mar 11;76(5):933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]