Abstract
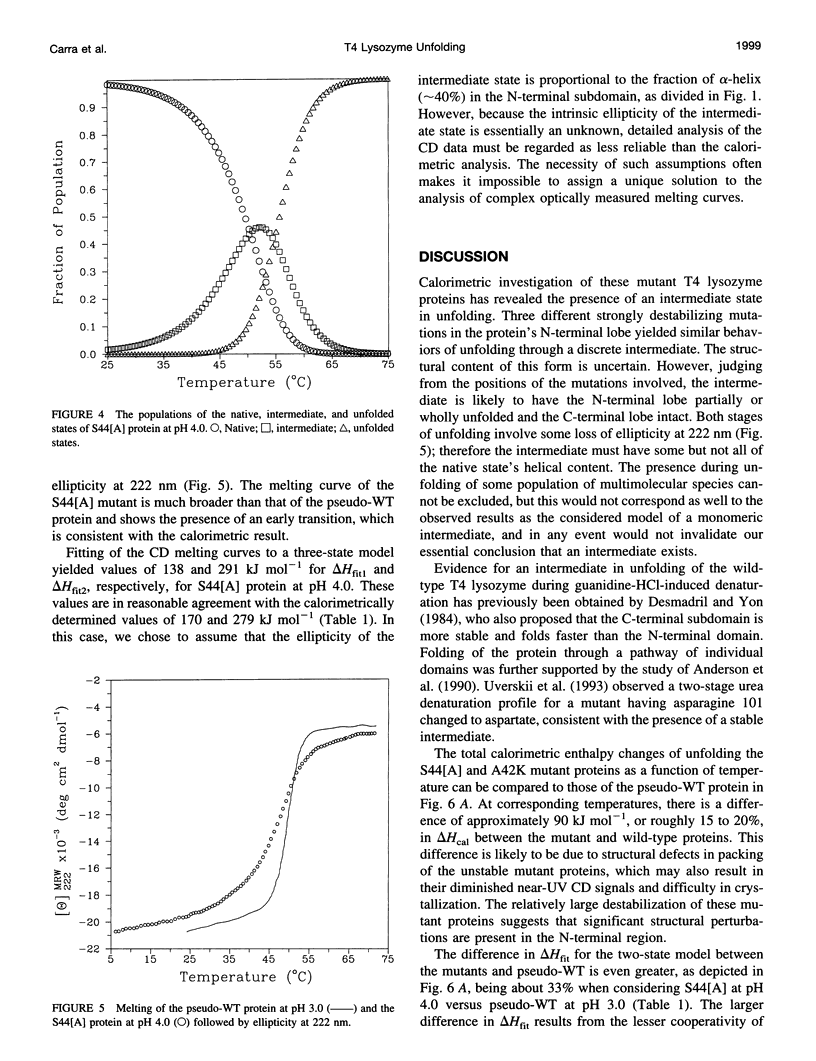
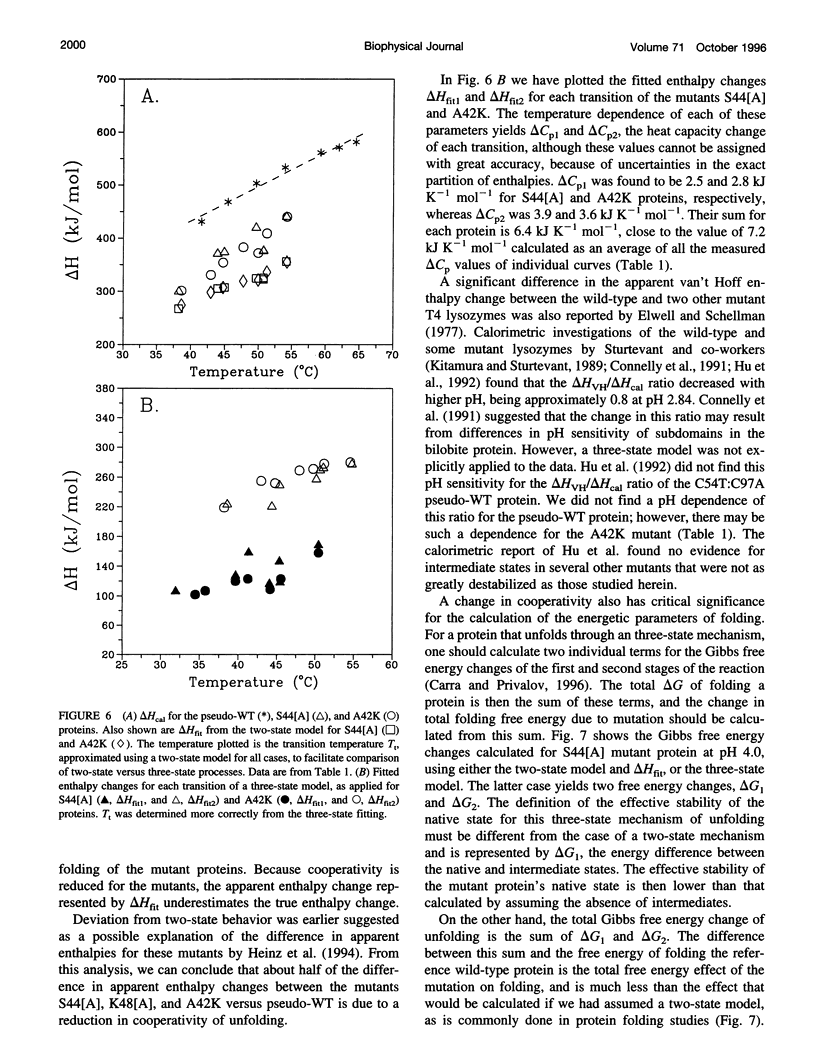
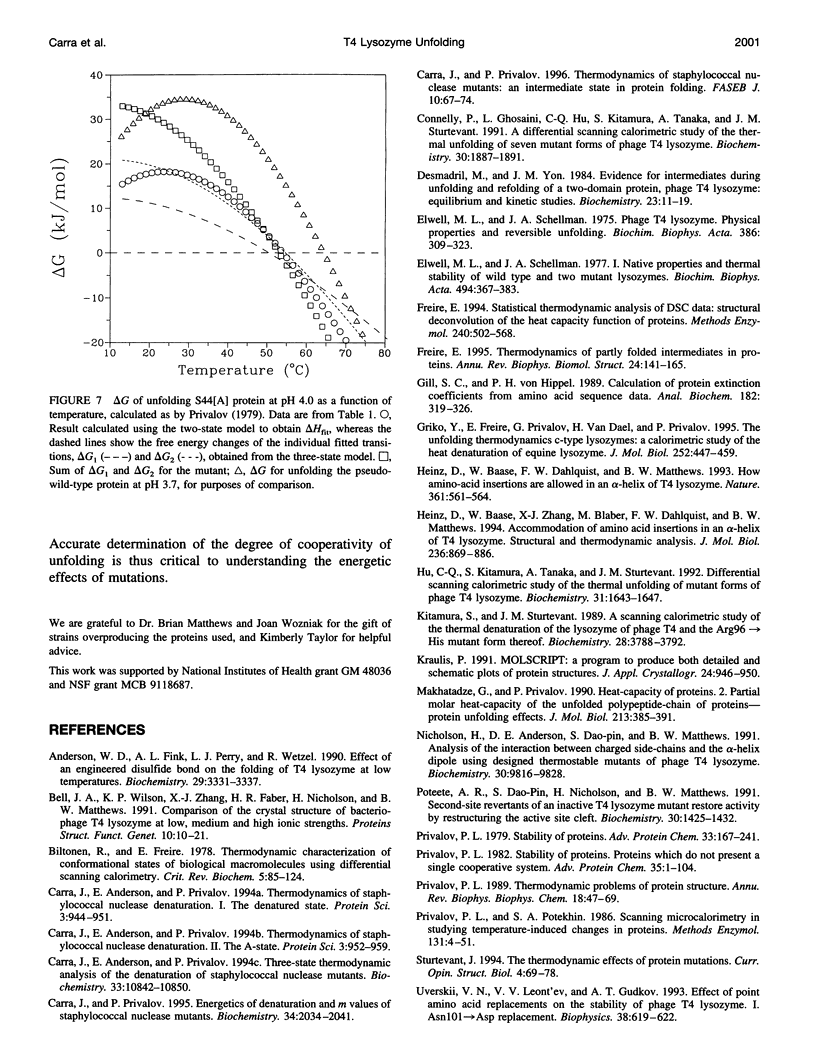
We investigated the folding of substantially destabilized mutant forms of T4 lysozyme using differential scanning calorimetry and circular dichroism measurements. Three mutations in an alpha-helix in the protein's N-terminal region, the alanine insertion mutations S44[A] and K48[A], and the substitution A42K had previously been observed to result in unexpectedly low apparent enthalpy changes of melting, compared to a pseudo-wild-type reference protein. The pseudo-wild-type reference protein thermally unfolds in an essentially two-state manner. However, we found that the unfolding of the three mutant proteins has reduced cooperativity, which partially explains their lower apparent enthalpy changes. A three-state unfolding model including a discrete intermediate is necessary to describe the melting of the mutant proteins. The reduction in cooperativity must be considered for accurate calculation of the energy changes of folding. Unfolding in two stages reflects the underlying two-subdomain structure of the lysozyme protein family.
Full text
PDF
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson W. D., Fink A. L., Perry L. J., Wetzel R. Effect of an engineered disulfide bond on the folding of T4 lysozyme at low temperatures. Biochemistry. 1990 Apr 3;29(13):3331–3337. doi: 10.1021/bi00465a026. [DOI] [PubMed] [Google Scholar]
- Bell J. A., Wilson K. P., Zhang X. J., Faber H. R., Nicholson H., Matthews B. W. Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths. Proteins. 1991;10(1):10–21. doi: 10.1002/prot.340100103. [DOI] [PubMed] [Google Scholar]
- Biltonen R. L., Freire E. Thermodynamic characterization of conformational states of biological macromolecules using differential scanning calorimetry. CRC Crit Rev Biochem. 1978;5(2):85–124. doi: 10.3109/10409237809177141. [DOI] [PubMed] [Google Scholar]
- Carra J. H., Anderson E. A., Privalov P. L. Thermodynamics of staphylococcal nuclease denaturation. I. The acid-denatured state. Protein Sci. 1994 Jun;3(6):944–951. doi: 10.1002/pro.5560030609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carra J. H., Anderson E. A., Privalov P. L. Thermodynamics of staphylococcal nuclease denaturation. II. The A-state. Protein Sci. 1994 Jun;3(6):952–959. doi: 10.1002/pro.5560030610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carra J. H., Anderson E. A., Privalov P. L. Three-state thermodynamic analysis of the denaturation of staphylococcal nuclease mutants. Biochemistry. 1994 Sep 6;33(35):10842–10850. doi: 10.1021/bi00201a035. [DOI] [PubMed] [Google Scholar]
- Carra J. H., Privalov P. L. Energetics of denaturation and m values of staphylococcal nuclease mutants. Biochemistry. 1995 Feb 14;34(6):2034–2041. doi: 10.1021/bi00006a025. [DOI] [PubMed] [Google Scholar]
- Carra J. H., Privalov P. L. Thermodynamics of denaturation of staphylococcal nuclease mutants: an intermediate state in protein folding. FASEB J. 1996 Jan;10(1):67–74. doi: 10.1096/fasebj.10.1.8566550. [DOI] [PubMed] [Google Scholar]
- Connelly P., Ghosaini L., Hu C. Q., Kitamura S., Tanaka A., Sturtevant J. M. A differential scanning calorimetric study of the thermal unfolding of seven mutant forms of phage T4 lysozyme. Biochemistry. 1991 Feb 19;30(7):1887–1891. doi: 10.1021/bi00221a022. [DOI] [PubMed] [Google Scholar]
- Desmadril M., Yon J. M. Evidence for intermediates during unfolding and refolding of a two-domain protein, phage T4 lysozyme: equilibrium and kinetic studies. Biochemistry. 1984 Jan 3;23(1):11–19. doi: 10.1021/bi00296a003. [DOI] [PubMed] [Google Scholar]
- Elwell M. L., Schellman J. A. Stability of phage T4 lysozymes. I. Native properties and thermal stability of wild type and two mutant lysozymes. Biochim Biophys Acta. 1977 Oct 26;494(2):367–383. doi: 10.1016/0005-2795(77)90166-0. [DOI] [PubMed] [Google Scholar]
- Elwell M., Schellman J. Phage T4 lysozyme. Physical properties and reversible unfolding. Biochim Biophys Acta. 1975 Mar 28;386(1):309–323. doi: 10.1016/0005-2795(75)90273-1. [DOI] [PubMed] [Google Scholar]
- Freire E. Statistical thermodynamic analysis of differential scanning calorimetry data: structural deconvolution of heat capacity function of proteins. Methods Enzymol. 1994;240:502–530. doi: 10.1016/s0076-6879(94)40062-8. [DOI] [PubMed] [Google Scholar]
- Freire E. Thermodynamics of partly folded intermediates in proteins. Annu Rev Biophys Biomol Struct. 1995;24:141–165. doi: 10.1146/annurev.bb.24.060195.001041. [DOI] [PubMed] [Google Scholar]
- Gill S. C., von Hippel P. H. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989 Nov 1;182(2):319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- Griko Y. V., Freire E., Privalov G., van Dael H., Privalov P. L. The unfolding thermodynamics of c-type lysozymes: a calorimetric study of the heat denaturation of equine lysozyme. J Mol Biol. 1995 Sep 29;252(4):447–459. doi: 10.1006/jmbi.1995.0510. [DOI] [PubMed] [Google Scholar]
- Heinz D. W., Baase W. A., Dahlquist F. W., Matthews B. W. How amino-acid insertions are allowed in an alpha-helix of T4 lysozyme. Nature. 1993 Feb 11;361(6412):561–564. doi: 10.1038/361561a0. [DOI] [PubMed] [Google Scholar]
- Heinz D. W., Baase W. A., Zhang X. J., Blaber M., Dahlquist F. W., Matthews B. W. Accommodation of amino acid insertions in an alpha-helix of T4 lysozyme. Structural and thermodynamic analysis. J Mol Biol. 1994 Feb 25;236(3):869–886. doi: 10.1006/jmbi.1994.1195. [DOI] [PubMed] [Google Scholar]
- Hu C. Q., Kitamura S., Tanaka A., Sturtevant J. M. Differential scanning calorimetric study of the thermal unfolding of mutant forms of phage T4 lysozyme. Biochemistry. 1992 Feb 18;31(6):1643–1647. doi: 10.1021/bi00121a009. [DOI] [PubMed] [Google Scholar]
- Kitamura S., Sturtevant J. M. A scanning calorimetric study of the thermal denaturation of the lysozyme of phage T4 and the Arg 96----His mutant form thereof. Biochemistry. 1989 May 2;28(9):3788–3792. doi: 10.1021/bi00435a024. [DOI] [PubMed] [Google Scholar]
- Nicholson H., Anderson D. E., Dao-pin S., Matthews B. W. Analysis of the interaction between charged side chains and the alpha-helix dipole using designed thermostable mutants of phage T4 lysozyme. Biochemistry. 1991 Oct 15;30(41):9816–9828. doi: 10.1021/bi00105a002. [DOI] [PubMed] [Google Scholar]
- Poteete A. R., Sun D. P., Nicholson H., Matthews B. W. Second-site revertants of an inactive T4 lysozyme mutant restore activity by restructuring the active site cleft. Biochemistry. 1991 Feb 5;30(5):1425–1432. doi: 10.1021/bi00219a037. [DOI] [PubMed] [Google Scholar]
- Privalov P. L., Makhatadze G. I. Heat capacity of proteins. II. Partial molar heat capacity of the unfolded polypeptide chain of proteins: protein unfolding effects. J Mol Biol. 1990 May 20;213(2):385–391. doi: 10.1016/S0022-2836(05)80198-6. [DOI] [PubMed] [Google Scholar]
- Privalov P. L., Potekhin S. A. Scanning microcalorimetry in studying temperature-induced changes in proteins. Methods Enzymol. 1986;131:4–51. doi: 10.1016/0076-6879(86)31033-4. [DOI] [PubMed] [Google Scholar]
- Privalov P. L. Stability of proteins. Proteins which do not present a single cooperative system. Adv Protein Chem. 1982;35:1–104. [PubMed] [Google Scholar]
- Privalov P. L. Stability of proteins: small globular proteins. Adv Protein Chem. 1979;33:167–241. doi: 10.1016/s0065-3233(08)60460-x. [DOI] [PubMed] [Google Scholar]
- Privalov P. L. Thermodynamic problems of protein structure. Annu Rev Biophys Biophys Chem. 1989;18:47–69. doi: 10.1146/annurev.bb.18.060189.000403. [DOI] [PubMed] [Google Scholar]



