Abstract
Bacteriophage P22 binds to its cell surface receptor, the repetitive O-antigen structure in Salmonella lipopolysaccharide, by its six homotrimeric tailspikes. Receptor binding by soluble tailspikes and the receptor-inactivating endorhamnosidase activity of the tailspike protein were studied using octa- and dodecasaccharides comprising two and three O-antigen repeats of Salmonella enteritidis and Salmonella typhimurium lipopolysaccharides. Wild-type tailspike protein and three mutants (D392N, D395N, and E359Q) with defective endorhamnosidase activity were used. Oligosaccharide binding to all three subunits, measured by a tryptophan fluorescence quench or by fluorescence depolarization of a coumarin label attached to the reducing end of the dodecasaccharide, occurs independently. At 10 degrees C, the binding affinities of all four proteins to oligosaccharides from both bacterial strains are identical within experimental error, and the binding constants for octa- and dodecasaccharides are 1 x 10(6) M(-1) and 2 x 10(6) M(-1), proving that two O-antigen repeats are sufficient for lipopolysaccharide recognition by the tailspike. Equilibration with the oligosaccharides occurs rapidly, but the endorhamnosidase produces only one cleavage every 100 s at 10 degrees C or about 2 min(-1) at the bacterial growth temperature. Thus, movement of virions in the lipopolysaccharide layer before DNA injection may involve the release and rebinding of individual tailspikes rather than hydrolysis of the O-antigen.
Full text
PDF

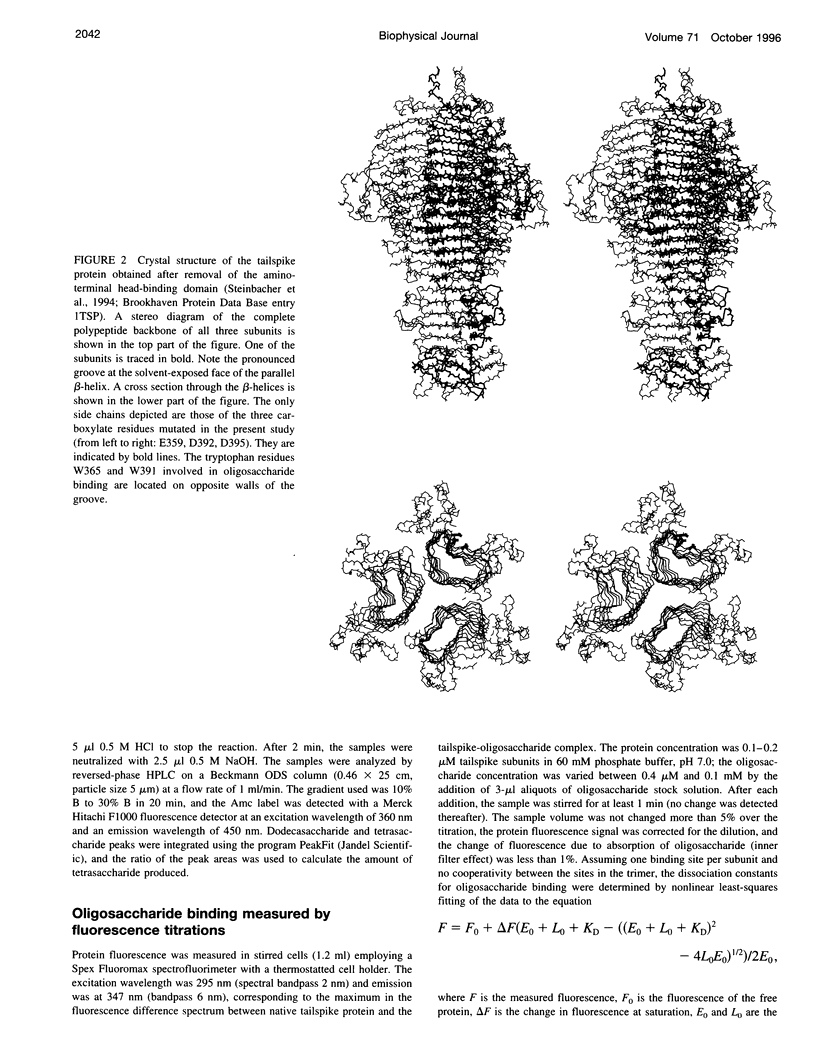

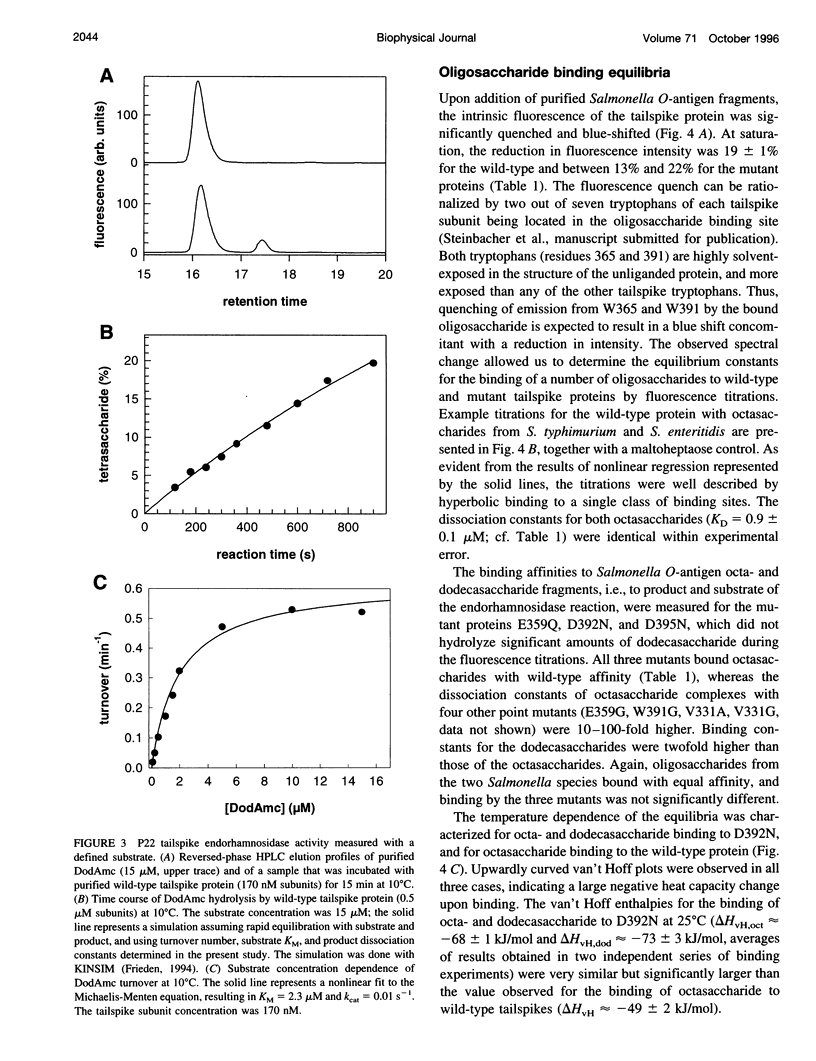
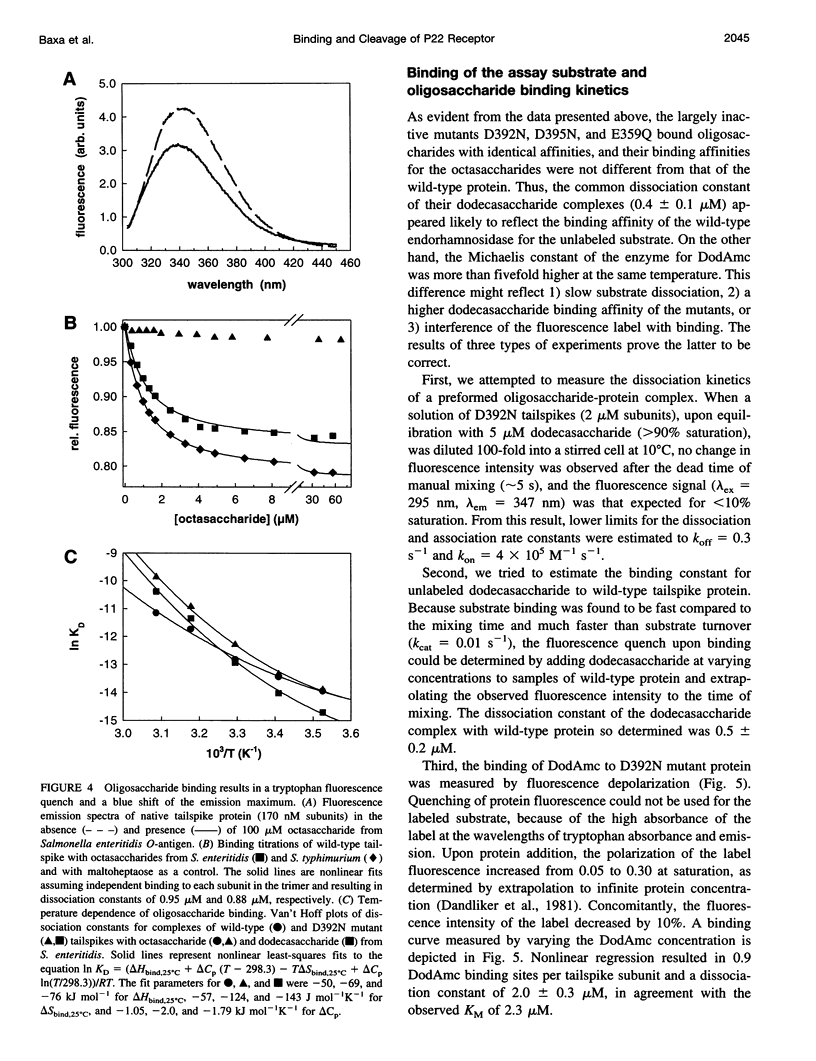
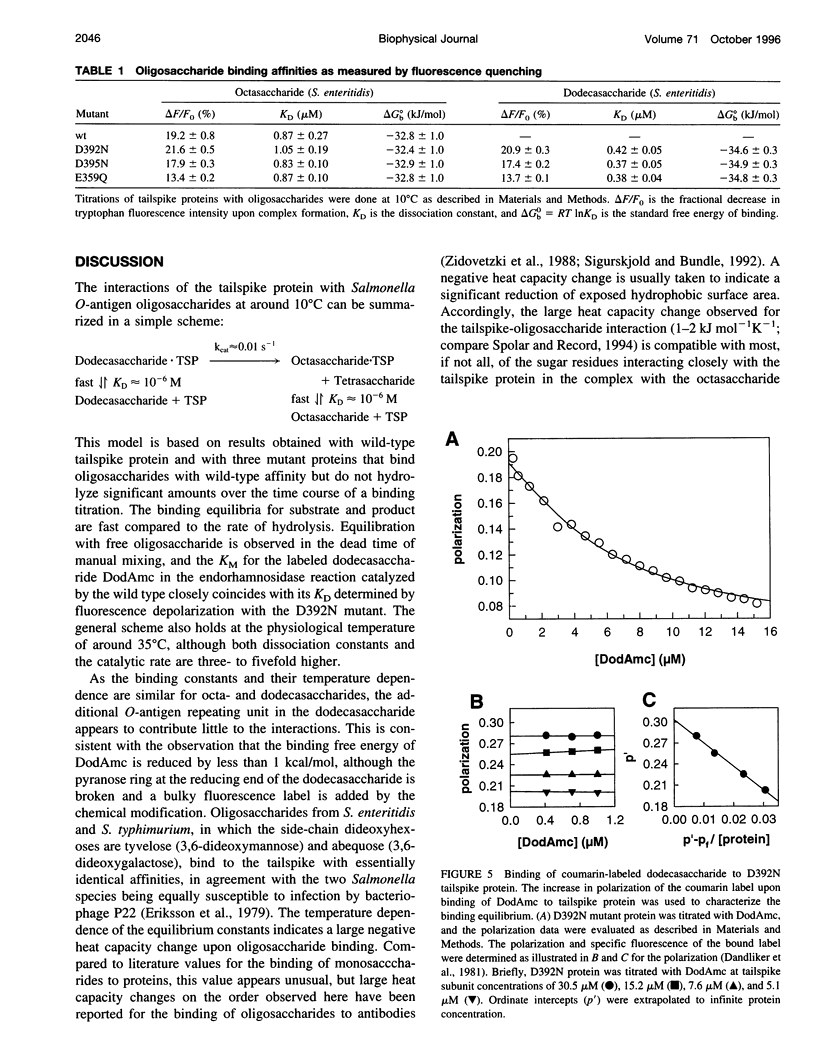


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bayer M. E., Takeda K., Uetake H. Effects of receptor destruction by Salmonella bacteriophages epsilon 15 and c341. Virology. 1980 Sep;105(2):328–337. doi: 10.1016/0042-6822(80)90034-3. [DOI] [PubMed] [Google Scholar]
- Berget P. B., Poteete A. R. Structure and functions of the bacteriophage P22 tail protein. J Virol. 1980 Apr;34(1):234–243. doi: 10.1128/jvi.34.1.234-243.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandliker W. B., Hsu M. L., Levin J., Rao B. R. Equilibrium and kinetic inhibition assays based upon fluorescence polarization. Methods Enzymol. 1981;74(Pt 100):3–28. doi: 10.1016/0076-6879(81)74003-5. [DOI] [PubMed] [Google Scholar]
- Danner M., Fuchs A., Miller S., Seckler R. Folding and assembly of phage P22 tailspike endorhamnosidase lacking the N-terminal, head-binding domain. Eur J Biochem. 1993 Aug 1;215(3):653–661. doi: 10.1111/j.1432-1033.1993.tb18076.x. [DOI] [PubMed] [Google Scholar]
- Eriksson U., Svenson S. B., Lönngren J., Lindberg A. A. Salmonella phage glycanases: substrate specificity of the phage P22 endo-rhamnosidase. J Gen Virol. 1979 Jun;43(3):503–511. doi: 10.1099/0022-1317-43-3-503. [DOI] [PubMed] [Google Scholar]
- Frieden C. Numerical integration of rate equations by computer: an update. Trends Biochem Sci. 1994 Apr;19(4):181–182. doi: 10.1016/0968-0004(94)90282-8. [DOI] [PubMed] [Google Scholar]
- Fuchs A., Seiderer C., Seckler R. In vitro folding pathway of phage P22 tailspike protein. Biochemistry. 1991 Jul 2;30(26):6598–6604. doi: 10.1021/bi00240a032. [DOI] [PubMed] [Google Scholar]
- Goldenberg D., King J. Trimeric intermediate in the in vivo folding and subunit assembly of the tail spike endorhamnosidase of bacteriophage P22. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3403–3407. doi: 10.1073/pnas.79.11.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrler G., Gross H. J., Imhof A., Brossmer R., Milks G., Paulson J. C. A synthetic sialic acid analogue is recognized by influenza C virus as a receptor determinant but is resistant to the receptor-destroying enzyme. J Biol Chem. 1992 Jun 25;267(18):12501–12505. [PubMed] [Google Scholar]
- Huberman K., Peluso R. W., Moscona A. Hemagglutinin-neuraminidase of human parainfluenza 3: role of the neuraminidase in the viral life cycle. Virology. 1995 Dec 1;214(1):294–300. doi: 10.1006/viro.1995.9925. [DOI] [PubMed] [Google Scholar]
- Israel J. V., Anderson T. F., Levine M. in vitro MORPHOGENESIS OF PHAGE P22 FROM HEADS AND BASE-PLATE PARTS. Proc Natl Acad Sci U S A. 1967 Feb;57(2):284–291. doi: 10.1073/pnas.57.2.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel V. A model for the adsorption of phage P22 to Salmonella typhimurium. J Gen Virol. 1978 Sep;40(3):669–673. doi: 10.1099/0022-1317-40-3-669. [DOI] [PubMed] [Google Scholar]
- Iwashita S., Kanegasaki S. Deacetylation reaction catalyzed by Salmonella phage c341 and its baseplate parts. J Biol Chem. 1976 Sep 10;251(17):5361–5365. [PubMed] [Google Scholar]
- Iwashita S., Kanegasaki S. Enzymic and molecular properties of base-plate parts of bacteriophage P22. Eur J Biochem. 1976 May 17;65(1):87–94. doi: 10.1111/j.1432-1033.1976.tb10392.x. [DOI] [PubMed] [Google Scholar]
- Kanaoka Y., Takahashi T., Nakayama H., Tanizawa K. New fluorogenic substrates for subtilisin. Chem Pharm Bull (Tokyo) 1985 Apr;33(4):1721–1724. doi: 10.1248/cpb.33.1721. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lentz T. L. The recognition event between virus and host cell receptor: a target for antiviral agents. J Gen Virol. 1990 Apr;71(Pt 4):751–766. doi: 10.1099/0022-1317-71-4-751. [DOI] [PubMed] [Google Scholar]
- Liu C., Eichelberger M. C., Compans R. W., Air G. M. Influenza type A virus neuraminidase does not play a role in viral entry, replication, assembly, or budding. J Virol. 1995 Feb;69(2):1099–1106. doi: 10.1128/jvi.69.2.1099-1106.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash C., Vijay I. K. A new fluorescent tag for labeling of saccharides. Anal Biochem. 1983 Jan;128(1):41–46. doi: 10.1016/0003-2697(83)90341-x. [DOI] [PubMed] [Google Scholar]
- Schwarz J. J., Berget P. B. Characterization of bacteriophage P22 tailspike mutant proteins with altered endorhamnosidase and capsid assembly activities. J Biol Chem. 1989 Nov 25;264(33):20112–20119. [PubMed] [Google Scholar]
- Seckler R., Fuchs A., King J., Jaenicke R. Reconstitution of the thermostable trimeric phage P22 tailspike protein from denatured chains in vitro. J Biol Chem. 1989 Jul 15;264(20):11750–11753. [PubMed] [Google Scholar]
- Sigurskjold B. W., Bundle D. R. Thermodynamics of oligosaccharide binding to a monoclonal antibody specific for a Salmonella O-antigen point to hydrophobic interactions in the binding site. J Biol Chem. 1992 Apr 25;267(12):8371–8376. [PubMed] [Google Scholar]
- Spolar R. S., Record M. T., Jr Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994 Feb 11;263(5148):777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- Steinbacher S., Seckler R., Miller S., Steipe B., Huber R., Reinemer P. Crystal structure of P22 tailspike protein: interdigitated subunits in a thermostable trimer. Science. 1994 Jul 15;265(5170):383–386. doi: 10.1126/science.8023158. [DOI] [PubMed] [Google Scholar]
- Svenson S. B., Lönngren J., Carlin N., Lindberg A. A. Salmonella bacteriophage glycanases: endorhamnosidases of Salmonella typhimurium bacteriophages. J Virol. 1979 Nov;32(2):583–592. doi: 10.1128/jvi.32.2.583-592.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withers S. G., Aebersold R. Approaches to labeling and identification of active site residues in glycosidases. Protein Sci. 1995 Mar;4(3):361–372. doi: 10.1002/pro.5560040302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zidovetzki R., Blatt Y., Schepers G., Pecht I. Thermodynamics of oligosaccharides binding to a dextran-specific monoclonal IgM. Mol Immunol. 1988 Apr;25(4):379–383. doi: 10.1016/0161-5890(88)90032-6. [DOI] [PubMed] [Google Scholar]



