Abstract
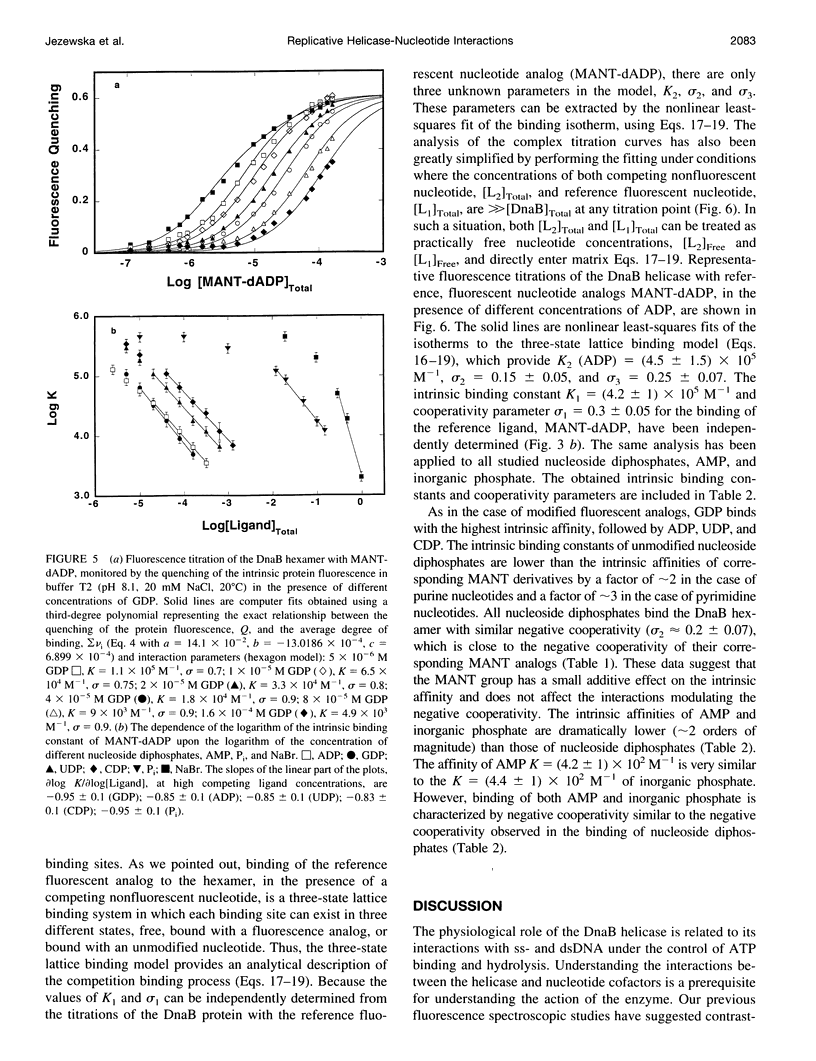
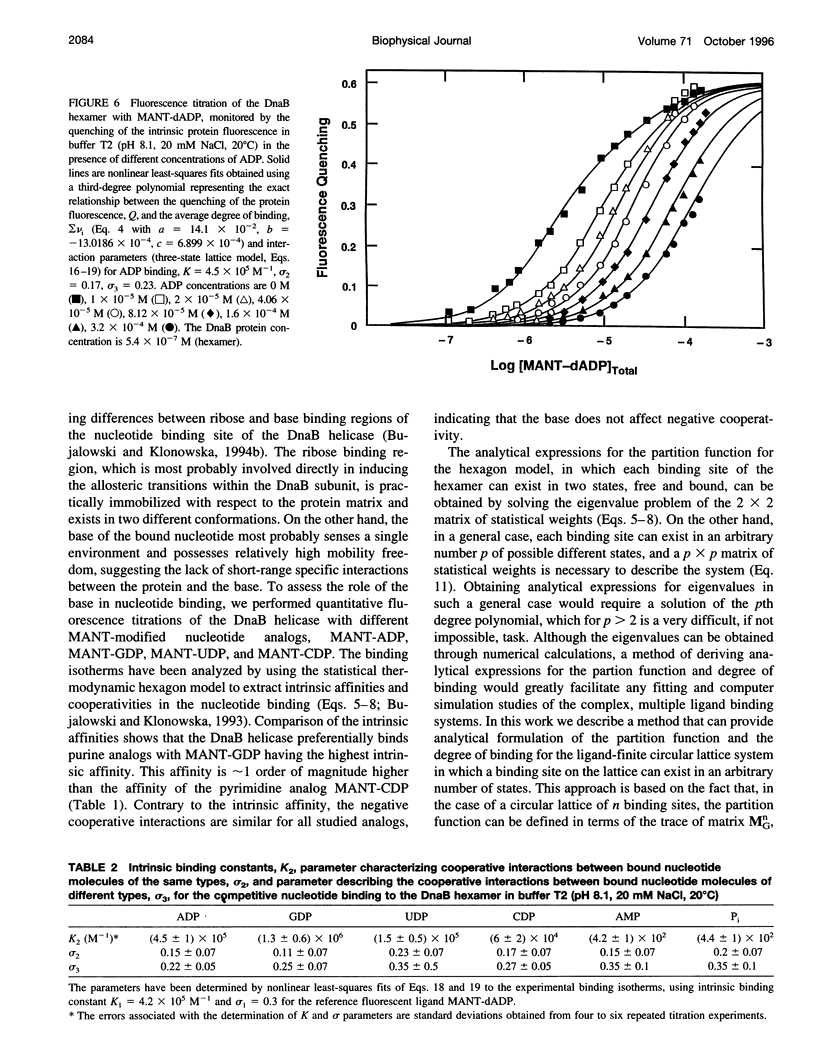
Interactions between the Escherichia coli primary replicative helicase DnaB protein and nucleotide cofactors have been studied using several fluorescent nucleotide analogs and unmodified nucleotides. The thermodynamically rigorous fluorescent titration technique has been used to obtain true binding isotherms, independently of the assumptions of any relationships between the observed quenching of protein fluorescence and the degree of nucleotide binding. Fluorescence titrations using several MANT derivatives of nucleoside diphosphates (MANT-ADP, 3',2'-O-(N-methylantraniloyl)adenosine-5'-diphosphate; MANT-GDP, 3',2'-O(N-methylantraniloyl)guanosine-5'-diphosphate; MANT-CDP, 3',2'-O-(N-methylantraniloyl)cytidine-5'-diphosphate; MANT-UDP, 3',2'-O-(N-methylantraniloyl)uridine-5'-diphosphate) have shown that the DnaB helicase has a preference for purine nucleotides. Binding of all modified nucleotides is characterized by similar negative cooperativity, indicating that negative cooperative interactions are base-independent. Thermodynamic parameters for the interactions of the unmodified nucleotides (ADP, GDP, CDP, and UDP) and inorganic phosphate (P(i)) have been obtained by using the competition titration approach. To analyze multiple ligand binding to a finite circular lattice, for a general case in which each lattice binding site can exist in different multiple states, we developed a matrix method approach to derive analytical expressions for the partition function and the average degree of binding for such cases. Application of the theory to competition titrations has allowed us to extract the intrinsic binding constants and cooperativity parameters for all unmodified ligands. This is the first quantitative estimate of affinities and the mechanisms of binding of different unmodified nucleotides and inorganic phosphate for a hexameric helicase. The intrinsic affinities of all of the studied ATP analogs are lower than the intrinsic affinities of the corresponding ADP analogs. The implications of these results for the mechanism of helicase action are discussed.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker T. A., Funnell B. E., Kornberg A. Helicase action of dnaB protein during replication from the Escherichia coli chromosomal origin in vitro. J Biol Chem. 1987 May 15;262(14):6877–6885. [PubMed] [Google Scholar]
- Biswas E. E., Biswas S. B., Bishop J. E. The dnaB protein of Escherichia coli: mechanism of nucleotide binding, hydrolysis, and modulation by dnaC protein. Biochemistry. 1986 Nov 18;25(23):7368–7374. doi: 10.1021/bi00371a019. [DOI] [PubMed] [Google Scholar]
- Bujalowski W., Jezewska M. J. Interactions of Escherichia coli primary replicative helicase DnaB protein with single-stranded DNA. The nucleic acid does not wrap around the protein hexamer. Biochemistry. 1995 Jul 11;34(27):8513–8519. doi: 10.1021/bi00027a001. [DOI] [PubMed] [Google Scholar]
- Bujalowski W., Klonowska M. M. Close proximity of tryptophan residues and ATP-binding site in Escherichia coli primary replicative helicase DnaB protein. Molecular topography of the enzyme. J Biol Chem. 1994 Dec 16;269(50):31359–31371. [PubMed] [Google Scholar]
- Bujalowski W., Klonowska M. M., Jezewska M. J. Oligomeric structure of Escherichia coli primary replicative helicase DnaB protein. J Biol Chem. 1994 Dec 16;269(50):31350–31358. [PubMed] [Google Scholar]
- Bujalowski W., Klonowska M. M. Negative cooperativity in the binding of nucleotides to Escherichia coli replicative helicase DnaB protein. Interactions with fluorescent nucleotide analogs. Biochemistry. 1993 Jun 8;32(22):5888–5900. doi: 10.1021/bi00073a023. [DOI] [PubMed] [Google Scholar]
- Bujalowski W., Klonowska M. M. Structural characteristics of the nucleotide-binding site of Escherichia coli primary replicative helicase DnaB protein. Studies with ribose and base-modified fluorescent nucleotide analogs. Biochemistry. 1994 Apr 19;33(15):4682–4694. doi: 10.1021/bi00181a028. [DOI] [PubMed] [Google Scholar]
- Cremo C. R., Neuron J. M., Yount R. G. Interaction of myosin subfragment 1 with fluorescent ribose-modified nucleotides. A comparison of vanadate trapping and SH1-SH2 cross-linking. Biochemistry. 1990 Apr 3;29(13):3309–3319. doi: 10.1021/bi00465a023. [DOI] [PubMed] [Google Scholar]
- Hiratsuka T. New ribose-modified fluorescent analogs of adenine and guanine nucleotides available as substrates for various enzymes. Biochim Biophys Acta. 1983 Feb 15;742(3):496–508. doi: 10.1016/0167-4838(83)90267-4. [DOI] [PubMed] [Google Scholar]
- Jencks W. P. The utilization of binding energy in coupled vectorial processes. Adv Enzymol Relat Areas Mol Biol. 1980;51:75–106. doi: 10.1002/9780470122969.ch2. [DOI] [PubMed] [Google Scholar]
- Jezewska M. J., Bujalowski W. A general method of analysis of ligand binding to competing macromolecules using the spectroscopic signal originating from a reference macromolecule. Application to Escherichia coli replicative helicase DnaB protein nucleic acid interactions. Biochemistry. 1996 Feb 20;35(7):2117–2128. doi: 10.1021/bi952344l. [DOI] [PubMed] [Google Scholar]
- Jezewska M. J., Bujalowski W. Global conformational transitions in Escherichia coli primary replicative helicase DnaB protein induced by ATP, ADP, and single-stranded DNA binding. Multiple conformational states of the helicase hexamer. J Biol Chem. 1996 Feb 23;271(8):4261–4265. doi: 10.1074/jbc.271.8.4261. [DOI] [PubMed] [Google Scholar]
- Jezewska M. J., Kim U. S., Bujalowski W. Binding of Escherichia coli primary replicative helicase DnaB protein to single-stranded DNA. Long-range allosteric conformational changes within the protein hexamer. Biochemistry. 1996 Feb 20;35(7):2129–2145. doi: 10.1021/bi952345d. [DOI] [PubMed] [Google Scholar]
- LeBowitz J. H., McMacken R. The Escherichia coli dnaB replication protein is a DNA helicase. J Biol Chem. 1986 Apr 5;261(10):4738–4748. [PubMed] [Google Scholar]
- Marians K. J. Prokaryotic DNA replication. Annu Rev Biochem. 1992;61:673–719. doi: 10.1146/annurev.bi.61.070192.003325. [DOI] [PubMed] [Google Scholar]
- Matson S. W., Kaiser-Rogers K. A. DNA helicases. Annu Rev Biochem. 1990;59:289–329. doi: 10.1146/annurev.bi.59.070190.001445. [DOI] [PubMed] [Google Scholar]
- McMacken R., Kornberg A. A multienzyme system for priming the replication of phiX174 viral DNA. J Biol Chem. 1978 May 10;253(9):3313–3319. [PubMed] [Google Scholar]
- Moore K. J., Lohman T. M. Kinetic mechanism of adenine nucleotide binding to and hydrolysis by the Escherichia coli Rep monomer. 2. Application of a kinetic competition approach. Biochemistry. 1994 Dec 6;33(48):14565–14578. doi: 10.1021/bi00252a024. [DOI] [PubMed] [Google Scholar]
- Reha-Krantz L. J., Hurwitz J. The dnaB gene product of Escherichia coli. II. Single stranded DNA-dependent ribonucleoside triphosphatase activity. J Biol Chem. 1978 Jun 10;253(11):4051–4057. [PubMed] [Google Scholar]
- San Martin M. C., Stamford N. P., Dammerova N., Dixon N. E., Carazo J. M. A structural model for the Escherichia coli DnaB helicase based on electron microscopy data. J Struct Biol. 1995 May-Jun;114(3):167–176. doi: 10.1006/jsbi.1995.1016. [DOI] [PubMed] [Google Scholar]
- Thompson C. J. Models for hemoglobin and allosteric enzymes. Biopolymers. 1968;6(8):1101–1118. doi: 10.1002/bip.1968.360060806. [DOI] [PubMed] [Google Scholar]
- Tougu K., Peng H., Marians K. J. Identification of a domain of Escherichia coli primase required for functional interaction with the DnaB helicase at the replication fork. J Biol Chem. 1994 Feb 11;269(6):4675–4682. [PubMed] [Google Scholar]
- Ueda K., McMacken R., Kornberg A. dnaB protein of Escherichia coli. Purification and role in the replication of phiX174 DNA. J Biol Chem. 1978 Jan 10;253(1):261–269. [PubMed] [Google Scholar]
- Wahle E., Lasken R. S., Kornberg A. The dnaB-dnaC replication protein complex of Escherichia coli. I. Formation and properties. J Biol Chem. 1989 Feb 15;264(5):2463–2468. [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner S., Wright M., Hurwitz J. Association of DNA-dependent and -independent ribonucleoside triphosphatase activities with dnaB gene product of Escherichia coli. Proc Natl Acad Sci U S A. 1974 Mar;71(3):783–787. doi: 10.1073/pnas.71.3.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Jezewska M. J., Bujalowski W., Egelman E. H. The hexameric E. coli DnaB helicase can exist in different Quaternary states. J Mol Biol. 1996 May 31;259(1):7–14. doi: 10.1006/jmbi.1996.0297. [DOI] [PubMed] [Google Scholar]


