Abstract
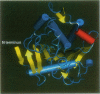
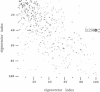
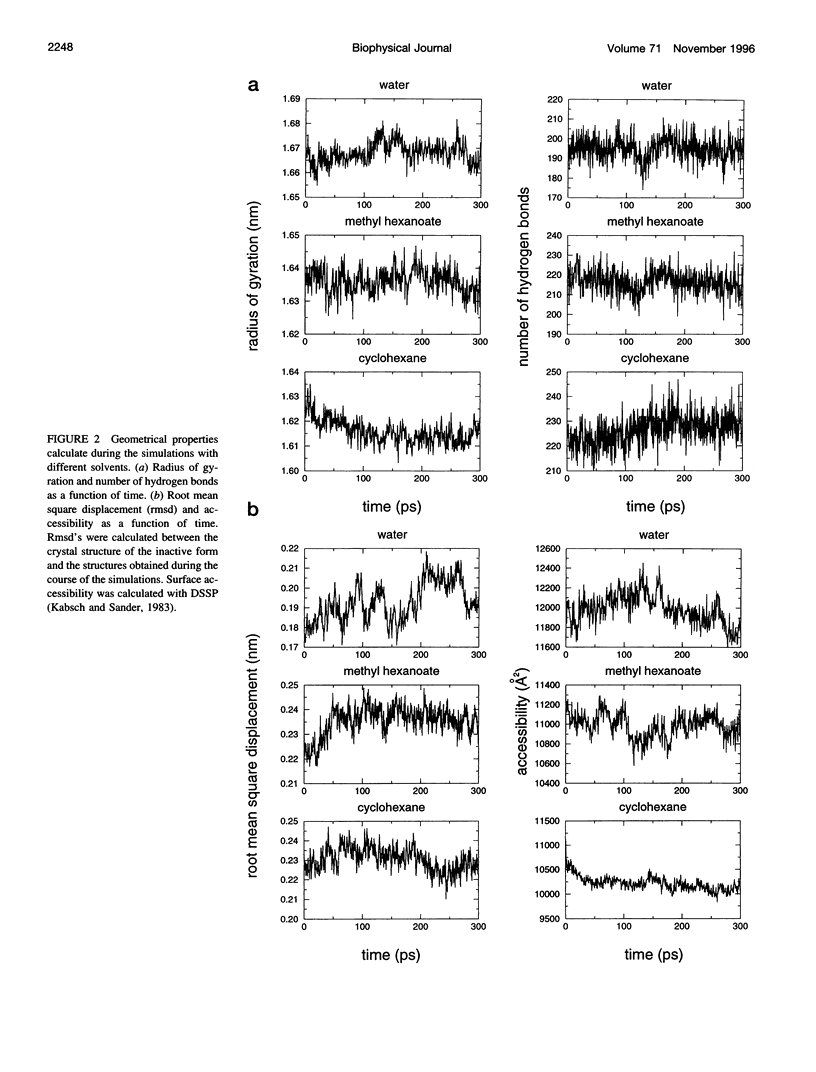
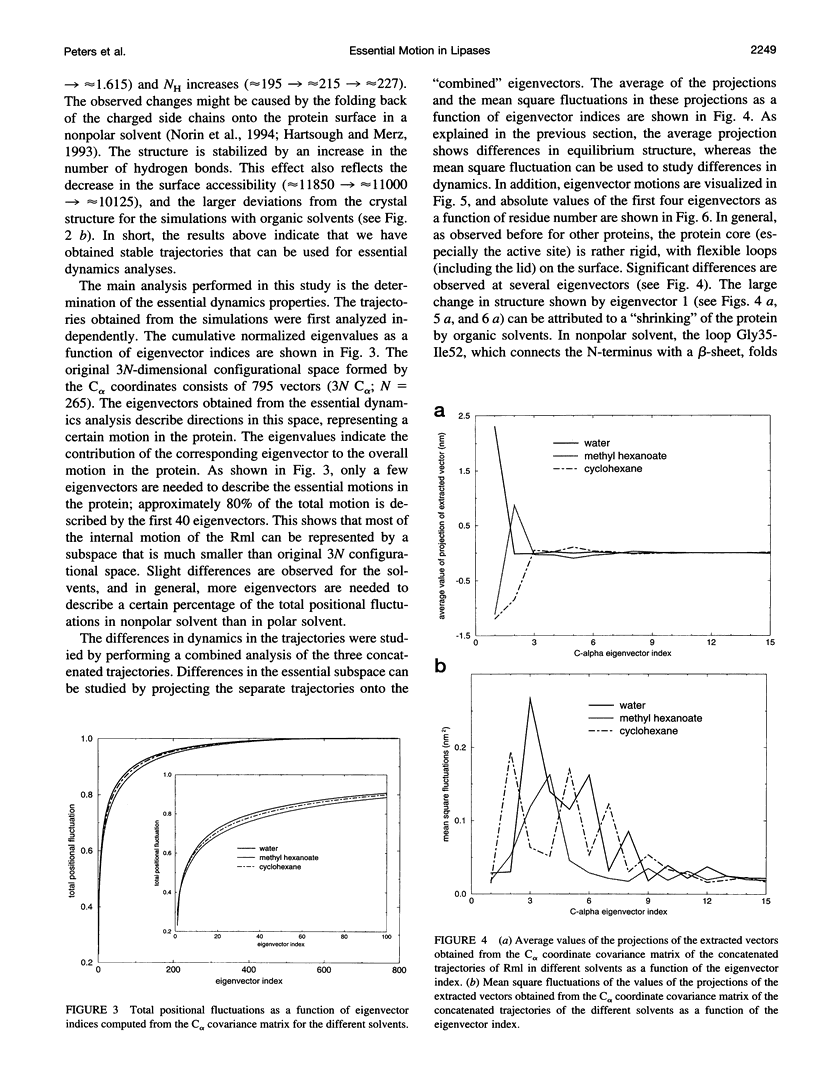
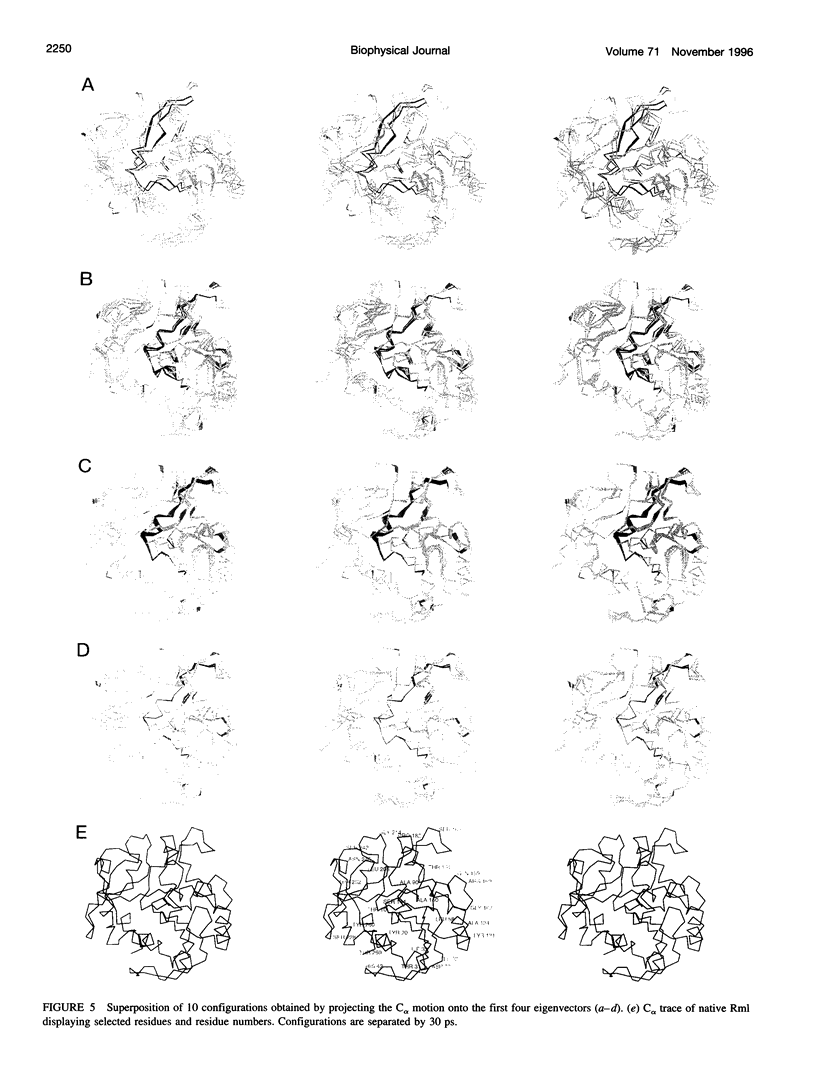
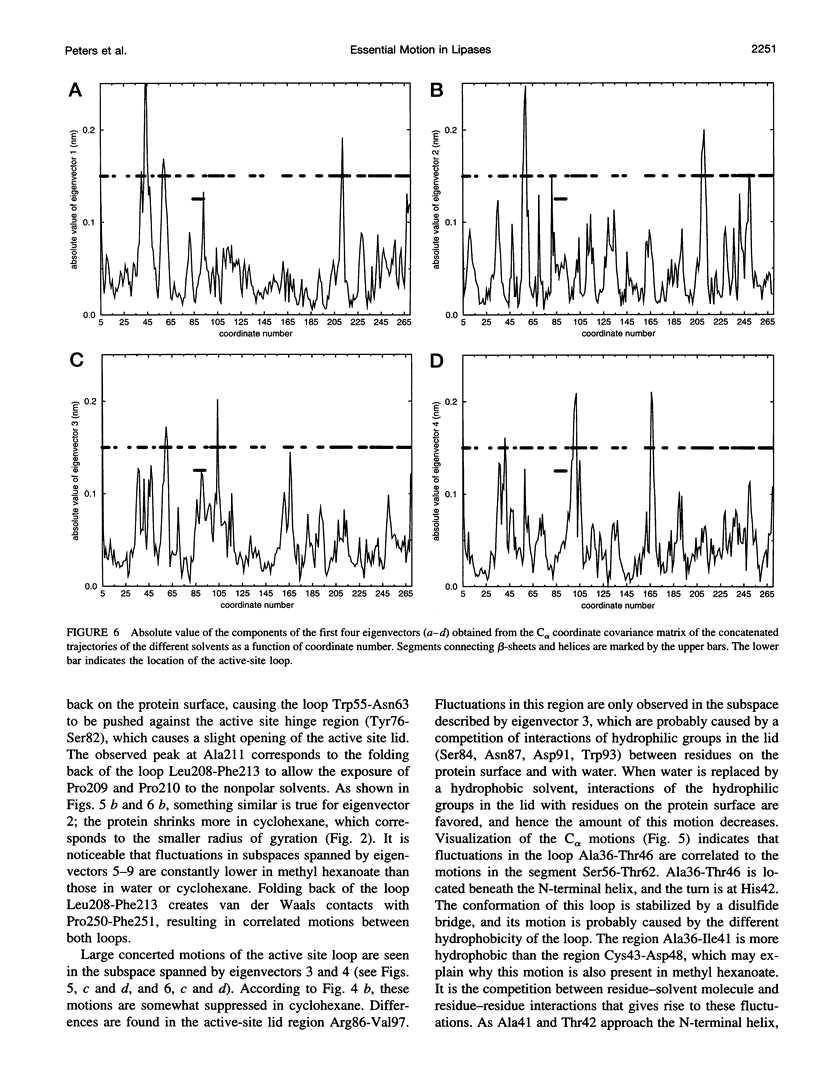
We have investigated the effect of different solvents on the dynamics of Rhizomucor miehei lipase. Molecular dynamics simulations were performed in water, methyl hexanoate, and cyclohexane. Analysis of the 400-ps trajectories showed that the solvent has a pronounced effect on the geometrical properties of the protein. The radius of gyration and total accessibility surface decrease in organic solvents, whereas the number of hydrogen bonds increases. The essential motions of the protein in different solvents can be described in a low-dimensional "essential subspace," and the dynamic behavior in this subspace correlates with the polarity of the solvent. Methyl hexanoate, which is a substrate for R. miehei lipase, significantly increases the fluctuations in the active-site loop. During the simulation, a methyl hexanoate entered the active-site groove. This observation provides insight into the possible docking mechanism of the substrate.
Full text
PDF


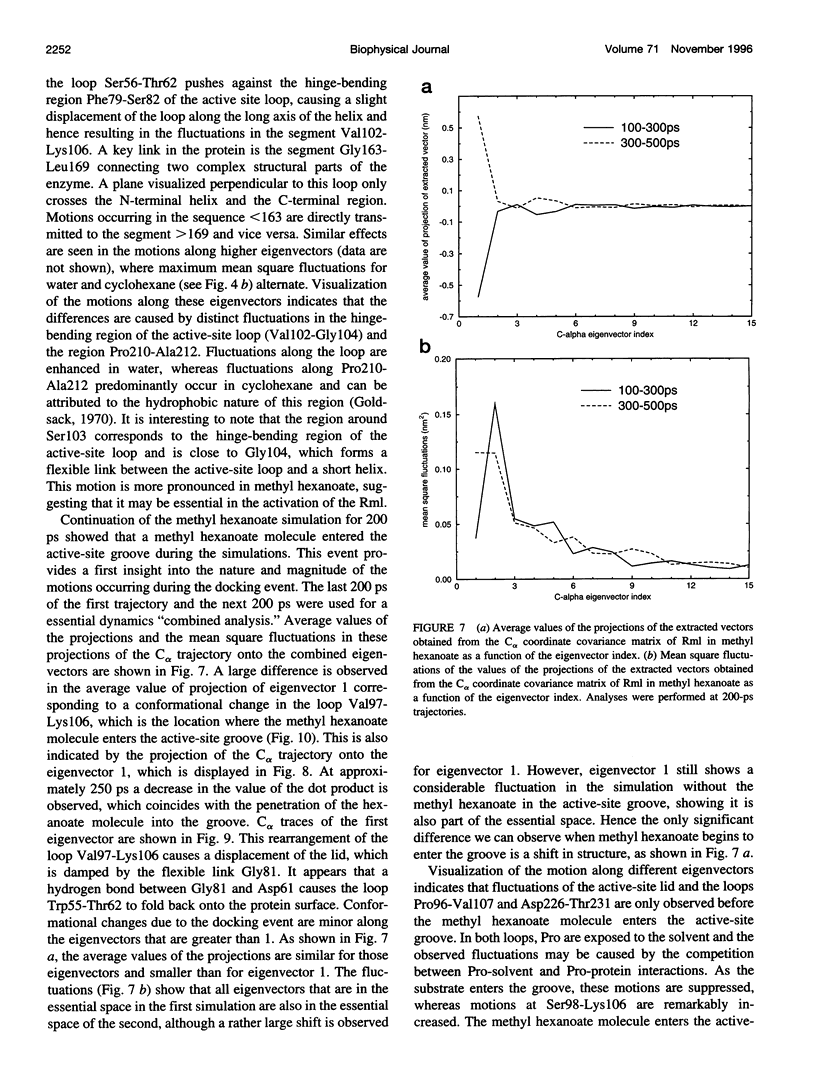
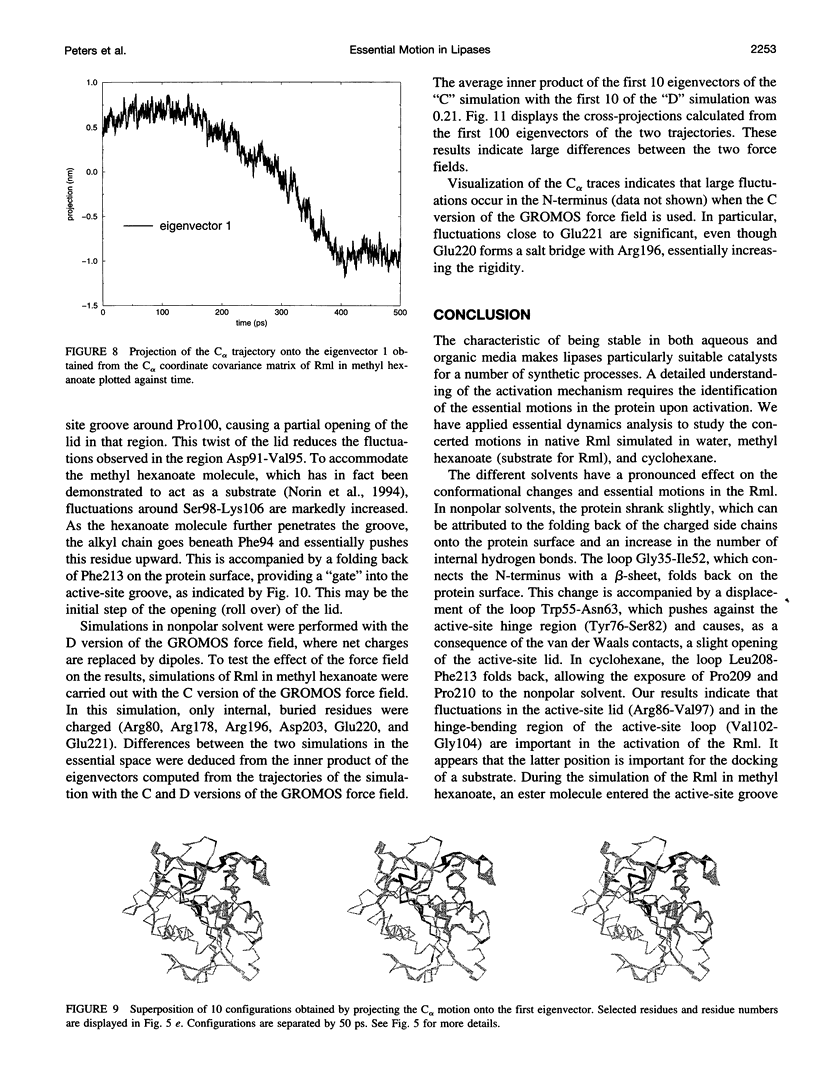
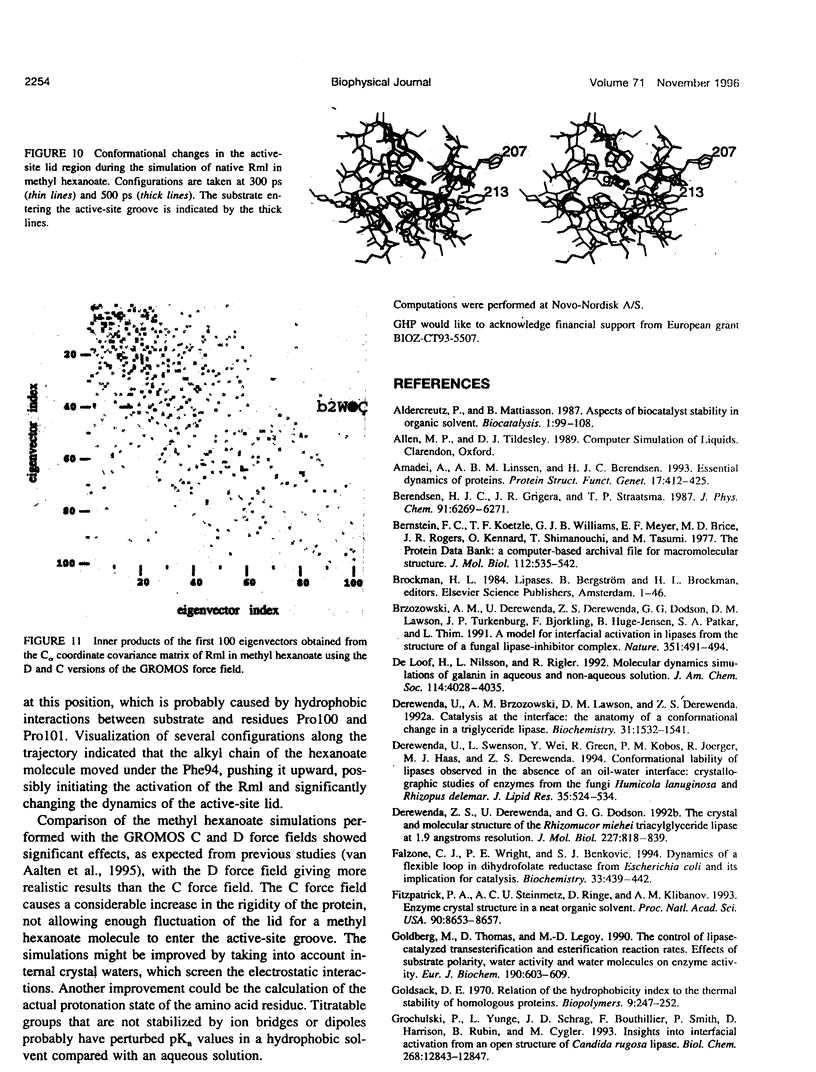

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amadei A., Linssen A. B., Berendsen H. J. Essential dynamics of proteins. Proteins. 1993 Dec;17(4):412–425. doi: 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Brzozowski A. M., Derewenda U., Derewenda Z. S., Dodson G. G., Lawson D. M., Turkenburg J. P., Bjorkling F., Huge-Jensen B., Patkar S. A., Thim L. A model for interfacial activation in lipases from the structure of a fungal lipase-inhibitor complex. Nature. 1991 Jun 6;351(6326):491–494. doi: 10.1038/351491a0. [DOI] [PubMed] [Google Scholar]
- Derewenda U., Brzozowski A. M., Lawson D. M., Derewenda Z. S. Catalysis at the interface: the anatomy of a conformational change in a triglyceride lipase. Biochemistry. 1992 Feb 11;31(5):1532–1541. doi: 10.1021/bi00120a034. [DOI] [PubMed] [Google Scholar]
- Derewenda U., Swenson L., Wei Y., Green R., Kobos P. M., Joerger R., Haas M. J., Derewenda Z. S. Conformational lability of lipases observed in the absence of an oil-water interface: crystallographic studies of enzymes from the fungi Humicola lanuginosa and Rhizopus delemar. J Lipid Res. 1994 Mar;35(3):524–534. [PubMed] [Google Scholar]
- Derewenda Z. S., Derewenda U., Dodson G. G. The crystal and molecular structure of the Rhizomucor miehei triacylglyceride lipase at 1.9 A resolution. J Mol Biol. 1992 Oct 5;227(3):818–839. doi: 10.1016/0022-2836(92)90225-9. [DOI] [PubMed] [Google Scholar]
- Falzone C. J., Wright P. E., Benkovic S. J. Dynamics of a flexible loop in dihydrofolate reductase from Escherichia coli and its implication for catalysis. Biochemistry. 1994 Jan 18;33(2):439–442. doi: 10.1021/bi00168a007. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick P. A., Steinmetz A. C., Ringe D., Klibanov A. M. Enzyme crystal structure in a neat organic solvent. Proc Natl Acad Sci U S A. 1993 Sep 15;90(18):8653–8657. doi: 10.1073/pnas.90.18.8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg M., Thomas D., Legoy M. D. The control of lipase-catalysed transesterification and esterification reaction rates. Effects of substrate polarity, water activity and water molecules on enzyme activity. Eur J Biochem. 1990 Jul 5;190(3):603–609. doi: 10.1111/j.1432-1033.1990.tb15615.x. [DOI] [PubMed] [Google Scholar]
- Goldsack D. E. Relation of the hydrophobicity index to the thermal stability of homologous proteins. Biopolymers. 1970 Feb;9(2):247–252. doi: 10.1002/bip.1970.360090209. [DOI] [PubMed] [Google Scholar]
- Grochulski P., Li Y., Schrag J. D., Bouthillier F., Smith P., Harrison D., Rubin B., Cygler M. Insights into interfacial activation from an open structure of Candida rugosa lipase. J Biol Chem. 1993 Jun 15;268(17):12843–12847. [PubMed] [Google Scholar]
- Gupta M. N. Enzyme function in organic solvents. Eur J Biochem. 1992 Jan 15;203(1-2):25–32. doi: 10.1111/j.1432-1033.1992.tb19823.x. [DOI] [PubMed] [Google Scholar]
- Jain M. K., Berg O. G. The kinetics of interfacial catalysis by phospholipase A2 and regulation of interfacial activation: hopping versus scooting. Biochim Biophys Acta. 1989 Apr 3;1002(2):127–156. doi: 10.1016/0005-2760(89)90281-6. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983 Dec;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kempner E. S. Movable lobes and flexible loops in proteins. Structural deformations that control biochemical activity. FEBS Lett. 1993 Jul 12;326(1-3):4–10. doi: 10.1016/0014-5793(93)81749-p. [DOI] [PubMed] [Google Scholar]
- Lautz J., Kessler H., van Gunsteren W. F., Weber H. P., Wenger R. M. On the dependence of molecular conformation on the type of solvent environment: a molecular dynamics study of cyclosporin A. Biopolymers. 1990 Oct-Nov;29(12-13):1669–1687. doi: 10.1002/bip.360291214. [DOI] [PubMed] [Google Scholar]
- Muderhwa J. M., Brockman H. L. Lateral lipid distribution is a major regulator of lipase activity. Implications for lipid-mediated signal transduction. J Biol Chem. 1992 Dec 5;267(34):24184–24192. [PubMed] [Google Scholar]
- Norin M., Haeffner F., Hult K., Edholm O. Molecular dynamics simulations of an enzyme surrounded by vacuum, water, or a hydrophobic solvent. Biophys J. 1994 Aug;67(2):548–559. doi: 10.1016/S0006-3495(94)80515-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters G. H., Olsen O. H., Svendsen A., Wade R. C. Theoretical investigation of the dynamics of the active site lid in Rhizomucor miehei lipase. Biophys J. 1996 Jul;71(1):119–129. doi: 10.1016/S0006-3495(96)79207-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters G. H., Toxvaerd S., Larsen N. B., Bjørnholm T., Schaumburg K., Kjaer K. Structure and dynamics of lipid monolayers: implications for enzyme catalysed lipolysis. Nat Struct Biol. 1995 May;2(5):395–401. doi: 10.1038/nsb0595-395. [DOI] [PubMed] [Google Scholar]
- Philippopoulos M., Xiang Y., Lim C. Identifying the mechanism of protein loop closure: a molecular dynamics simulation of the Bacillus stearothermophilus LDH loop in solution. Protein Eng. 1995 Jun;8(6):565–573. doi: 10.1093/protein/8.6.565. [DOI] [PubMed] [Google Scholar]
- Piéroni G., Gargouri Y., Sarda L., Verger R. Interactions of lipases with lipid monolayers. Facts and questions. Adv Colloid Interface Sci. 1990 Sep;32(4):341–378. doi: 10.1016/0001-8686(90)80023-s. [DOI] [PubMed] [Google Scholar]
- Ransac S., Rivière C., Soulié J. M., Gancet C., Verger R., de Haas G. H. Competitive inhibition of lipolytic enzymes. I. A kinetic model applicable to water-insoluble competitive inhibitors. Biochim Biophys Acta. 1990 Mar 12;1043(1):57–66. doi: 10.1016/0005-2760(90)90110-j. [DOI] [PubMed] [Google Scholar]
- Schrag J. D., Cygler M. 1.8 A refined structure of the lipase from Geotrichum candidum. J Mol Biol. 1993 Mar 20;230(2):575–591. doi: 10.1006/jmbi.1993.1171. [DOI] [PubMed] [Google Scholar]
- Thuren T. A model for the molecular mechanism of interfacial activation of phospholipase A2 supporting the substrate theory. FEBS Lett. 1988 Feb 29;229(1):95–99. doi: 10.1016/0014-5793(88)80805-6. [DOI] [PubMed] [Google Scholar]
- Vriend G. WHAT IF: a molecular modeling and drug design program. J Mol Graph. 1990 Mar;8(1):52-6, 29. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- Wade R. C., Davis M. E., Luty B. A., Madura J. D., McCammon J. A. Gating of the active site of triose phosphate isomerase: Brownian dynamics simulations of flexible peptide loops in the enzyme. Biophys J. 1993 Jan;64(1):9–15. doi: 10.1016/S0006-3495(93)81335-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. C., McDermott A. E. Dynamics of the flexible loop of triosephosphate isomerase: the loop motion is not ligand gated. Biochemistry. 1995 Jul 4;34(26):8309–8319. doi: 10.1021/bi00026a012. [DOI] [PubMed] [Google Scholar]
- Zaks A., Klibanov A. M. Enzymatic catalysis in nonaqueous solvents. J Biol Chem. 1988 Mar 5;263(7):3194–3201. [PubMed] [Google Scholar]
- van Aalten D. M., Amadei A., Bywater R., Findlay J. B., Berendsen H. J., Sander C., Stouten P. F. A comparison of structural and dynamic properties of different simulation methods applied to SH3. Biophys J. 1996 Feb;70(2):684–692. doi: 10.1016/S0006-3495(96)79608-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Aalten D. M., Amadei A., Linssen A. B., Eijsink V. G., Vriend G., Berendsen H. J. The essential dynamics of thermolysin: confirmation of the hinge-bending motion and comparison of simulations in vacuum and water. Proteins. 1995 May;22(1):45–54. doi: 10.1002/prot.340220107. [DOI] [PubMed] [Google Scholar]
- van Aalten D. M., Findlay J. B., Amadei A., Berendsen H. J. Essential dynamics of the cellular retinol-binding protein--evidence for ligand-induced conformational changes. Protein Eng. 1995 Nov;8(11):1129–1135. doi: 10.1093/protein/8.11.1129. [DOI] [PubMed] [Google Scholar]