Abstract
Two hundred eight of 648 (32%) diarrheal stool samples collected from hospitalized children under 5 years of age during a 3-year period (1999, 2000, and 2002) in the city of Salvador, in the state of Bahia, Brazil, were rotavirus positive. One hundred sixty-four of 208 (78.8%) rotavirus-positive samples had genotype G9 specificity, predominantly in association with P[8]. Other specificities detected were G1 (12.0%) and G4 (1.4%). Viruses with G2, G3, or P[4] specificity were not detected. Rotavirus genotype G9 predominated during each of the three seasons studied; it represented 89.2% of rotavirus strains detected in 1999, 85.3% in 2000, and 74.5% in 2002. G1 viruses (the globally most common G type) have a unique epidemiological characteristic of maintaining predominance during multiple consecutive rotavirus seasons. We have shown in this study for the first time that the G9 viruses also have a similar epidemiological characteristic, albeit for a shorter period of surveillance. The next generation of rotavirus vaccines will need to provide adequate protection against disease caused by G9 viruses.
Rotaviruses, members of the Reoviridae family, comprise seven groups (groups A to G). Group A rotaviruses have been well established as the single most important cause of severe acute gastroenteritis in infants and young children in both developed and developing countries. Each year, they are estimated to cause 111 million episodes of diarrhea requiring only home care, 25 million clinic visits, 2 million hospitalizations, and 325,000 to 592,000 deaths (median of 440,000 deaths) in children of <5 years of age (30). The medical, social, and economic burdens associated with rotavirus disease make it imperative to develop and implement a safe and effective rotavirus vaccine. Because serotype-specific protection appears to play an important role against rotavirus disease (for reviews, see references 16, 19, and 21), rotavirus typing surveillance studies remain essential.
An infectious group A rotavirus particle possesses 11 double-stranded RNA (dsRNA) segments surrounded by three concentric protein layers. The outer capsid consists of VP7 and VP4 proteins that carry independent neutralization and protective antigens (6); antibodies to either protein can confer resistance to virulent rotavirus in a type-specific manner in experimental animals (for a review, see reference 20). Thus far, 15 VP7 (G) and 24 VP4 (P) rotavirus genotypes have been described (20, 25, 26, 29, 33). Global epidemiological surveys have consistently shown that four G types (G1 to G4) and two P types (P[4] and P[8]) represent more than 90% of strains of clinical importance globally (9, 22, 38). A more recent epidemiological analysis of rotavirus G- and P-type distribution, however, has demonstrated (i) the global distribution and clinical importance of type G9 and (ii) the regional distribution and clinical importance of strains with G5, G8, or P[6] specificity (38). Hence, efforts have been made or are under consideration to develop human rotavirus candidate vaccines that bear such G and/or P types of epidemiological importance (for reviews, see references 21 and 38).
Rotavirus strain surveillance studies have been conducted annually since 1982 in various locations in Brazil (1, 2, 24, 28, 34, 36, 39, 44). However, a systematic rotavirus surveillance study has never been conducted in the city of Salvador, located in the state of Bahia in Northeast Brazil. It is the third largest city in the country. Here we report the results of a 3-year surveillance study of rotavirus strains among hospitalized infants and young children (<5 years old) with diarrhea in the city of Salvador, Brazil.
MATERIALS AND METHODS
Specimens.
Six hundred forty-eight stool samples were collected from children under 5 years of age hospitalized with acute diarrhea at the Hospital Centro Pediátrico Hosanah de Oliveira in the city of Salvador, Bahia, Brazil, during 1999 (n = 170), 2000 (n = 120), and 2002 (n = 358). All children were from poor areas with low levels of sanitation and socioeconomic backgrounds. The samples were screened for rotavirus by using polyacrylamide gel electrophoresis (PAGE) analysis only (13). No other viral pathogens were sought.
RNA extraction and typing PCR.
Rotavirus dsRNA was extracted from stools by (i) the phenol-chloroform method (10) for PAGE analysis or (ii) the Trizol method (Invitrogen, Carlsbad, CA) for typing PCR. Three different sets (i.e., the H2 pool, C pool, and A pool) (37) of type-specific primers for the VP7 gene and one set of type-specific primers for the VP4 gene (8) were used. The H2 pool contained G1-, G2-, G3-, G4-, G8-, and G9-specific primers (aBT1, aCT2, S3, S4, S8, and aFT9, respectively) (10; 41); the C pool consisted of G1-, G2-, G3-, G4-, and G9-specific primers (9T1-1, 9T1-2, 9T-3P, 9T-4, and 9T-9B, respectively) (5); and the A pool contained G5-, G6-, G8-, G10-, and G11-specific primers (FT5, DT6, HT8, G-10, and BT11, respectively) (11, 17). P typing was performed with P[4]-, P[6]-, P[8]-, P[9]-, and P[10]-specific primers (1T-1, 2T-1, 3T-1, 4T-1, and 5T-1, respectively) (8). The dsRNA samples were subjected to 1 cycle of reverse transcription (RT) (42°C, 45 min) followed by 30 cycles of PCR. Each PCR cycle for G or P genotyping contained steps of 1 min at 94°C, 2 min at 50°C, and 3 min at 72°C. The PCR products were analyzed by agarose gel electrophoresis and visualized by staining with ethidium bromide. Rotavirus strains Wa (G1P[8]), K8 (G1P [9]), DS-1 (G2P[4]), P (G3P[8]), RRV (G3P[3]), ST-3 (G4P[6]), OSU (G5P[7]), UK (G6P[5]), 69M (G8P[10]), WI-61 (G9P[8]), B223 (G10P[11]), and YM (G11P[7]) were used as control viruses.
Sequence alignment and phylogenetic analysis.
Three G9 samples from Salvador (BA206, isolated in 1999; BA201, isolated in 2000; and BA202, isolated in 2002) were randomly selected. The nucleotide sequence of the VP7 gene of each virus was determined with a BigDye terminator cycle sequencing kit (Applied Biosystems, Foster City, CA) and an ABI PRISM 310 automated DNA sequencer, and the sequence alignment and phylogenetic analysis were done by using the CLUSTAL W algorithm (42) of the MegAlign program in the DNASTAR software package (Madison, WI). The VP7 gene sequences of selected other G9 strains were obtained from GenBank.
Analysis of the dsRNA of selected rotavirus strains by PAGE.
The electrophoretic migration patterns of dsRNAs of the six G9 rotavirus strains (BA187, BA190, BA200, BA201, BA202, and BA206) from Salvador were compared to those of three G9 rotavirus strains detected in Rio de Janeiro, Brazil, in 1997 (strain R44) and 1999 (strains R143 and R168) (36). The extracted genomic dsRNAs were analyzed on a standard discontinuous 10% acrylamide slab gel with a 3.5% acrylamide stacking gel. The dsRNA bands were visualized by staining the gel with silver nitrate (13).
Determination of rotavirus subgroup (VP6) specificity by ELISA.
The VP6 subgroup specificities of selected G9 rotavirus strains (BA201, BA202, BA206, R44, R143, and R168) were determined by enzyme-linked immunosorbent assay (ELISA) using monoclonal antibodies specific for rotavirus subgroup I (SG-I) (antibody 266/60) or SG-II (antibody 631/9) that were generated at the National Institutes of Health (12). Briefly, the ELISA plate was coated with an anti-human rotavirus goat serum (930) diluted in carbonate buffer, pH 9.6. One hundred microliters/well of 10% stool suspension or infected cell culture lysates was added to four wells, and the plate was incubated overnight at 4°C. One hundred microliters/well of subgroup-specific (SG-I or SG-II) monoclonal antibody was added to two of the four wells for each sample, and the plate was incubated at 37°C for 1 h, followed by the addition of anti-mouse immunoglobulin G conjugated with horseradish peroxidase and incubation at 37°C for 1 h. The substrate was added to each well, and the optical density values were measured at 450 nm. Rotavirus strains DS-1 (SG-I) and Wa (SG-II) were used as controls.
Nucleotide sequence accession numbers.
The nucleotide sequences obtained in this study were deposited in GenBank under the accession numbers AY695809 (BA201), AY695810 (BA202), and AY695811 (BA206).
RESULTS
Two hundred eight of 648 (32%) diarrheal stool samples collected from hospitalized children under 5 years of age during a 3-year period (1999, 2000, and 2002) in the city of Salvador, located in the state of Bahia, Brazil, were rotavirus positive by PAGE analysis. G genotypes were successfully determined for 196 of the 208 (94.2%) rotavirus-positive samples; the VP7 gene of 12 strains (5.8%) could not be amplified. The distribution of G types detected in this study is shown in Table 1. One hundred sixty-four of the 208 samples (78.8%) were identified as rotavirus type G9, mostly in association with P[8] specificity. Other specificities detected were G1 (12.0%) and G4 (1.4%); G2 or G3 viruses were not detected. In 1999, G9 represented 89.2% (33/37) of the rotavirus strains analyzed; in 2000, it represented 85.3% (29/34), and in 2002, 74.5% (102/137) (Table 1). P genotypes were successfully determined for 179 of the 208 (86.1%) rotavirus-positive samples. For 29 samples (13.9%), despite several attempts, the RT-PCR VP4 gene product of such samples could not be obtained and, therefore, we did not have the opportunity to genotype by multiplex PCR and/or sequence analysis. Since all positive samples after first-round RT-PCR were successfully P genotyped, we did not use P-typing primers different from those described in Materials and Methods. All but one sample (99.4%) were typed P[8]; one sample from 2002 was typed as P[6]. Type P[4], which is the second most common P type worldwide, was not detected. Both G and P genotypes were determined for 172 (82.7%) samples. The distribution of G-P genotype combinations is shown in Table 2. G9P[8] (74.5%) was predominant throughout the 3-year surveillance period, followed by G1P[8] (5.8%) and G4P[8] (1.4%).
TABLE 1.
Distribution of human group A rotavirus G genotypes detected in this study
| Year | No. of rotavirus isolates | No. (%) of VP7 genotypea: |
No. (%) of: |
|||
|---|---|---|---|---|---|---|
| G1 | G4 | G9 | Nontypeable strains | Mixed G infections | ||
| 1999 | 37 | 2 (5.4) | 1 (2.7) | 33 (89.2) | 1 (2.7) | 0 (0) |
| 2000 | 34 | 1 (2.9) | 1 (2.9) | 29 (85.3) | 3 (8.8) | 0 (0) |
| 2002 | 137 | 22 (16.1) | 1 (0.7) | 102 (74.5) | 8 (5.8) | 4b (2.9) |
| Total | 208 | 25 (12.0) | 3 (1.4) | 164 (78.8) | 12 (5.8) | 4 (2.0) |
Mixed rotavirus infections are not included in the G1, G4, or G9 total.
All four mixed infections were caused by G1 + G9 strains.
TABLE 2.
G-P genotype combinations detected among hospitalized children in Salvador, Bahia, Brazil, during 1999, 2000, and 2002a
| G-P combination | No. of isolates detected in: |
Total no. (%) | ||
|---|---|---|---|---|
| 1999 | 2000 | 2002 | ||
| G1P[8] | 1 | 0 | 11 | 12 (5.8) |
| G1P?b | 1 | 1 | 11 | 13 (6.2) |
| G4P[8] | 1 | 1 | 1 | 3 (1.4) |
| G9P[8] | 33 | 27 | 95 | 155 (74.5) |
| G9P? | 0 | 2 | 7 | 9 (4.3) |
| G?P[8] | 0 | 2 | 4 | 6 (2.9) |
| G?P[6] | 0 | 0 | 1 | 1 (0.5) |
| G1 + G9P[8]c | 0 | 0 | 2 | 2 (1.0) |
| G1 + G9P? | 0 | 0 | 2 | 2 (1.0) |
| G?P? | 1 | 1 | 3 | 5 (2.4) |
| Total | 37 | 34 | 137 | 208 (100) |
Numbers and combinations shown in bold indicate that G9 strains were counted.
?, genotype not determined.
Mixed infection.
In this study, we used three different sets (i.e., H2 pool, C pool, and A pool) of type-specific primers in order to identify the VP7 specificities of the isolates, as previously suggested (37). The H2 pool contains primers specific for G1 to G4, G8, and G9 (10, 41). The G9-specific primer in this pool was chosen from the VP7 gene sequence of the prototype G9 WI61 strain, which belongs to lineage 1. The C pool contains primers specific for G1 to G4 and G9 (5). The G9 primer in this pool was chosen from the VP7 gene sequence of the Indian G9 116E strain, which belongs to lineage 2. The A pool contains primers specific for G5, G6, G8, G10, and G11 (11, 17). It has been reported that (i) G9 strains in lineage 1 are correctly typed as G9 with both primer pools (H2 and C), (ii) lineage 2 strains can be mistyped as negative or G9/G4 with pool H2 and correctly typed as G9 with pool C, and (iii) G9 strains belonging to lineage 3 can be mistyped as negative, G4, or G9/G4 with pool H2 and correctly typed as G9 with pool C (37). In the present study, 59.8% (98/164) of the G9 strains were mistyped as G4 or a mixture of G4 and G9 by the H2 pool. Furthermore, 40.2% (66/164) strains that gave negative results with the H2 pool were correctly genotyped as G9 by the C pool.
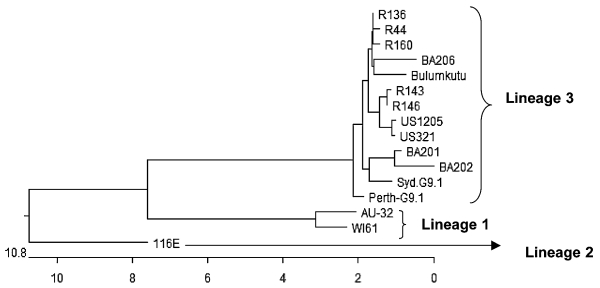
Sequence and phylogenetic analyses of the VP7 gene of three Salvador G9 strains (one strain from each year of the study) demonstrated that (i) each belonged to VP7 lineage 3 and (ii) each was closely related molecularly to the G9 strains previously isolated in Rio de Janeiro (Fig. 1; Table 3). The VP7 sequences of Salvador G9 strains isolated in 1999 (BA206) and 2000 (BA201) were shown to be more closely related phylogenetically to those of Rio de Janeiro G9 strains isolated between 1997 and 1999 than to the Salvador G9 strain (BA202) isolated in 2002 (Fig. 1).
FIG. 1.
Phylogenetic analysis of the VP7 gene nucleotide sequences of human rotavirus G9 strains. The dendrogram was constructed by the CLUSTAL W algorithm of the MegAlign program in the DNASTAR software package (Madison, WI). The units at the bottom of the tree indicate the distances between sequence pairs. Rotavirus strains and their corresponding GenBank accession numbers: R143, AF274969; R146, AF274970; R160, AF274971; R44, AF438227; R136, AF438228; Bulumkutu, AF359358; BA206, AY695809; BA202, AY695810; BA201, AY695811; US321, AJ250275; US1205, AF060487; Syd.G9.1, AY307093; Perth-G9.1, AY307094; AU-32, AB045372; WI61, AB180969; 116E, L14072.
TABLE 3.
Percentages of VP7 gene nucleotide identity among rotavirus G9 strains isolated from different locations and three rotavirus G9 isolates from Salvador, Bahia, Brazil
| G9 rotavirus strain | VP7 gene lineage | % Nucleotide identity with strain: |
||
|---|---|---|---|---|
| BA206/Brazil/BA-99 | BA201/Brazil/BA-00 | BA202/Brazil/BA-02 | ||
| R44/Brazil/RJ-97 | 3 | 98.5 | 98.4 | 97.3 |
| R136/Brazil/RJ-98 | 3 | 98.7 | 98.7 | 97.6 |
| R143/Brazil/RJ-99 | 3 | 98.0 | 98.2 | 97.3 |
| R146/Brazil/RJ-99 | 3 | 98.1 | 98.3 | 97.4 |
| R160/Brazil/RJ-99 | 3 | 98.5 | 98.5 | 97.4 |
| Bulumkutu/Nigeria-99/00 | 3 | 97.9 | 97.6 | 96.7 |
| Perth-G9.1/Australia-99 | 3 | 97.8 | 98.3 | 97.2 |
| Syd.G9.1/Australia-99 | 3 | 97.8 | 98.4 | 97.6 |
| US321/USA-97/98 | 3 | 98.0 | 98.2 | 97.1 |
| US1205/USA-96/97 | 3 | 97.8 | 97.9 | 97.2 |
| AU-32/Japan-85 | 1 | 87.5 | 87.5 | 86.3 |
| WI61/USA-83 | 1 | 87.8 | 88.2 | 87.4 |
| 116E/India-90 | 2 | 86.7 | 87.3 | 86.7 |
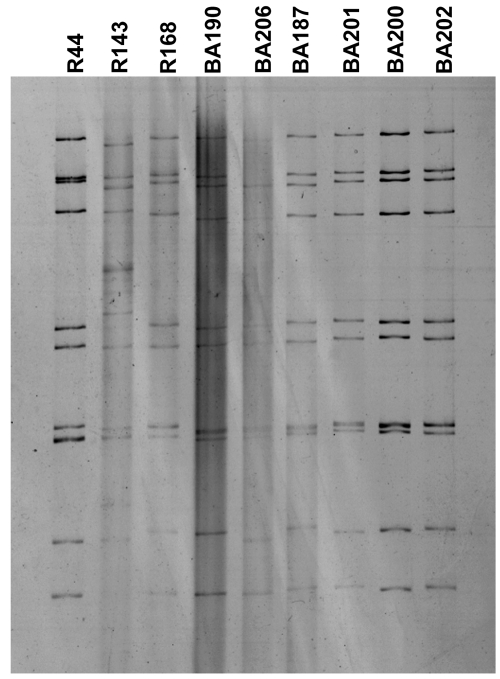
Each of the six G9P[8] rotavirus strains (BA190, BA206, BA187, BA201, BA200, and BA202) from Salvador analyzed by PAGE exhibited a “long” RNA pattern which was similar to that of the G9P[8] strain (R168) isolated in Rio de Janeiro (Fig. 2). The G9P[6] strain (R143) isolated in Rio de Janeiro demonstrated a “short” pattern, whereas the G9P[9] Rio de Janeiro strain R44 displayed a “long” pattern distinct from those exhibited by Salvador and Rio de Janeiro G9P[8] strains (Fig. 2).
FIG. 2.
Electrophoretic migration patterns of genomic RNAs of G9 rotavirus strains detected in Brazil between 1997 and 2002. Strain R44 (G9P[9]) was detected in Rio de Janeiro in 1997, and strains R143 (G9P[4]) and R168 (G9P[8]) were detected in Rio de Janeiro in 1999. Strains BA190 and BA206 were detected in Salvador in 1999, strains BA187 and BA201 in 2000, and strains BA200 and BA202 in 2002. All samples from Salvador possess G9P[8] specificity.
Five G9 samples (three [BA201, BA202, and BA206] from Salvador and two [R44 and R168] from Rio de Janeiro) that were tested for VP6 were shown to bear subgroup II specificity. A sixth sample, R143 from Rio de Janeiro, was shown to possess subgroup I specificity (data not shown).
DISCUSSION
Genomic point mutations and accumulations have been proposed to be one of three mechanisms of rotavirus evolution in nature, the other two being genome rearrangements and genetic reassortments (43). Previous studies have reported either a failure to genotype or the mistyping of contemporary G9 rotavirus strains, due to the accumulation of point mutations at the primer binding site (18, 27, 37) or the occurrence of antigenic variability (4). In this study, we used two different sets of primer pools (H2 pool and C pool), each of which contained a distinct G9 type-specific primer (aFT9 in the H2 pool and 9T-9B in the C pool), in order to identify correctly the VP7 specificity of the isolates, as previously suggested (37). The results were consistent with those previously described (37) and demonstrated a great VP7 gene polymorphism among G9 rotavirus strains. These findings reinforce the need for constant monitoring and updating of methods used for rotavirus typing (7, 18).
The first description of rotavirus type G9 was reported in the United States in the early 1980s (3). Soon after its detection, it disappeared for about a decade but then reemerged in the mid-1990s. Today, type G9 is globally the fourth most common rotavirus genotype detected in humans (38). Sequence analysis has demonstrated the existence of at least three distinct VP7 lineages among the rotavirus G9 strains (37). Most of the G9 viruses, which are prevalent around the world today, belong to lineage 3. In Brazil, rotavirus G9 was first detected in 1997 in Rio de Janeiro, located approximately 1,037 miles southeast of Salvador (1, 36). The G9 strains detected in that study (36) exhibited the same PCR profiles against H2 and C primer pools as those observed in the present study. It is of interest that the VP7 sequences of strains BA206 and BA201, isolated in 1999 and 2000, respectively, are more closely related phylogenetically to those of G9 strains isolated in Rio de Janeiro between 1997 and 1999 than to those of strain BA202 detected in 2002 in Salvador. In addition, each of the six G9P[8] rotavirus strains from Salvador analyzed by PAGE exhibited a “long” RNA pattern very similar to that of strain R168 isolated in Rio de Janeiro in 1999. Furthermore, three strains (BA201, BA202, and BA206) of the six Salvador G9P[8] strains used for VP7 gene sequence analysis and the R168 strain were shown to bear subgroup II specificity. Such observations suggest that (i) the G9 viruses may have spread in Brazil from the Southeast (Rio de Janeiro) to the Northeast (Salvador) and (ii) some level of VP7 gene sequence divergence, albeit subtle, occurred during the spread. In the United States, seasonal rotavirus activity has also been documented to occur in a sequential manner, beginning first in the Southwest from October through December and ending in the Northeast in April or May (45).
Recently, rotavirus G9 was reported as the predominant strain during two rotavirus seasons (2000-2001 and 2002-2003) among hospitalized children in Belgium (31). Rotavirus G1 was predominant during the intervening season of 2001-2002. In the current study, we describe the predominance of rotavirus G9 in Salvador, Bahia, Brazil, during at least three seasons (including two consecutive seasons, 1999 and 2000, with a percentage over 85%), among children hospitalized for rotavirus diarrhea. Unfortunately, we could not analyze samples from the 2001 season because they were not collected. G1 viruses (the globally most common G type) have a unique epidemiological characteristic of maintaining predominance during multiple consecutive rotavirus seasons (for a review, see reference 38). We have shown for the first time that G9 viruses also have a similar epidemiological characteristic, albeit for a shorter period of surveillance.
The age distribution of children infected with rotavirus G9 was between 1 month and 3 years old, the same as that of children infected with G1 strains. Unfortunately, we do not have more-detailed information on the severity of the infections; however, all children that participated in this study were hospitalized because of severe diarrhea. Rotavirus G9 has also been reported as the causative agent of severe diarrhea among hospitalized children in other countries (31, 40). It is interesting that a recent 2-year rotavirus surveillance study in Sweden reported (35) that G9 viruses were the dominant type (42.9%) detected in adults with acute diarrhea, followed by G1 viruses (30.6%), whereas in contrast, during the same period of surveillance, G1 viruses were detected most (72%) in children with acute diarrhea, followed by G9 viruses (20%). The authors speculated that these G9 virus infections occurred independently of preexisting immunity (35).
The importance of serotype-specific protection against rotavirus disease is still under discussion (for reviews, see references 16, 19, and 21); however, many vaccine candidates that have been developed are designed to provide antigenic coverage for globally or regionally important G and/or P types (for a review, see reference 38). Considering that rotavirus serotype G9 has emerged as the fourth most common rotavirus serotype detected in humans around the world (38), future rotavirus vaccine candidates may need to include a G9 component. However, since there is a pronounced diverse VP7 gene polymorphism among G9 rotavirus strains (23, 32, 37), it is important to determine if a G9 rotavirus vaccine candidate belonging to one VP7 lineage can provide protection against disease caused by G9 viruses belonging to a different VP7 lineage(s). Hoshino et al. (14) analyzed the relationship of the phylogenetic lineages to the neutralization specificities of various G9 strains and reported that antibodies raised against lineage 1 G9 strains, represented by the earliest strains recovered in the United States and Japan, efficiently neutralized the contemporary G9 strains from lineages 2 and 3; however, antibodies against the lineage 3 G9 strains did not efficiently neutralize strains belonging to lineage 1. Thus, it was concluded that the lineage 1 strain should be included as the G9 representative in a multivalent rotavirus vaccine, since it provided the broadest available reactivity to the extant G9 serotype (15). It is important, therefore, to maintain surveillance of rotavirus strains, particularly in developing areas where mixed rotavirus infections have been shown to promote reassortment with high frequency. Such events may lead to the appearance of new strains or new variants that could escape immune protection induced by an outdated vaccine.
Acknowledgments
We thank Alice Costa da Silva, Jerri Ross, and Ronald Jones for their technical assistance and Albert Kapikian for continuing support of our work.
This study was partially supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), and Fundação de Amparo à Pesquisa do Estado da Bahia (FAPESB), Brazil.
REFERENCES
- 1.Araújo, I. T., M. S. R. Ferreira, A. M. Fialho, R. M. Assis, C. M. Cruz, M. Rocha, and J. P. G. Leite. 2001. Rotavirus genotypes P[4]G9, P[6]G9, and P[8]G9 in hospitalized children with acute gastroenteritis in Rio de Janeiro, Brazil. J. Clin. Microbiol. 39:1999-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carmona, R. C. C., M. C. S. T. Timenetsky, F. F. da Silva, and C. F. H. Granato. 2004. Characterization of rotavirus strains from hospitalized and outpatient children with acute diarrhoea in São Paulo, Brazil. J. Med. Virol. 74:166-172. [DOI] [PubMed] [Google Scholar]
- 3.Clark, H. F., Y. Hoshino, L. M. Bell, J. Groff, G. Hess, P. Bachman, and P. A. Offit. 1987. Rotavirus isolate WI61 representing a presumptive new human serotype. J. Clin. Microbiol. 25:1757-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coulson, B. S., J. R. Gentsch, B. K. Das, M. K. Bhan, and R. I. Glass. 1999. Comparison of enzyme immunoassay and reverse transcriptase PCR for identification of serotype G9 rotaviruses. J. Clin. Microbiol. 37:3187-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das, B. K., J. R. Gentsch, H. G. Cicirello, P. A. Woods, A. Gupta, M. Ramachandran, R. Kumar, M. K. Bhan, and R. I. Glass. 1994. Characterization of rotavirus strains from newborns in New Delhi, India. J. Clin. Microbiol. 32:1820-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Estes, M. K. 2001. Rotaviruses and their replication, p. 1747-1785. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol 2. Lippincott, Williams & Wilkins, Philadelphia, Pa.
- 7.Fischer, T. K., and J. R. Gentsch. 2004. Rotavirus typing methods and algorithms. Rev. Med. Virol. 14:71-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gentsch, J. R., R. I. Glass, P. Woods, V. Gouvea, M. Gorziglia, J. Flores, B. K. Das, and M. K. Bhan. 1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 30:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gentsch, J. R., P. A. Woods, M. Rmachandran, B. K. Das, J. P. Leite, A. Alfieri, R. Kumar, M. K. Bhan, and R. I. Glass. 1996. Review of G and P typing results from a global collection of rotavirus strains: implications for vaccine development. J. Infect. Dis. 174(Suppl. 1):S30-S36. [DOI] [PubMed] [Google Scholar]
- 10.Gouvea, V., R. I. Glass, P. Woods, K. Taniguchi, H. F. Clark, B. Forrester, and Z.-Y. Fang. 1990. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 28:276-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gouvea, V., N. Santos, and M. C. Timenetsky. 1994. Identification of bovine and porcine rotavirus G types by PCR. J. Clin. Microbiol. 32:1338-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenberg, H., V. McAuliffe, J. Valdesuso, R. Wyatt, J. Flores, A. Kalica, Y. Hoshino, and N. Singh. 1983. Serological analysis of the subgroup protein of rotavirus, using monoclonal antibodies. Infect. Immun. 39:91-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herring, A. J., N. F. Inglis, C. K. Ojeh, D. R. Snodgrass, and J. D. Menzies. 1982. Rapid diagnosis of rotavirus infection by direct detection of viral nucleic acid in silver-stained polyacrylamide gels. J. Clin. Microbiol. 16:473-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoshino, Y., R. W. Jones, J. Ross, S. Honma, N. Santos, J. R. Gentsch, and A. Z. Kapikian. 2004. Rotavirus serotype G9 strains belonging to VP7 gene phylogenetic sequence lineage 1 may be more suitable for serotype G9 vaccine candidates than those belonging to lineage 2 or 3. J. Virol. 78:7795-7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoshino, Y., R. W. Jones, J. Ross, and A. Z. Kapikian. 2003. Construction and characterization of rhesus monkey rotavirus (MMU18006)- or bovine rotavirus (UK)-based serotype G5, G8, G9 or G10 single VP7 gene substitution reassortant candidate vaccines. Vaccine 21:3003-3010. [DOI] [PubMed] [Google Scholar]
- 16.Hoshino, Y., and A. Z. Kapikian. 2000. Rotavirus serotypes: classification and importance in epidemiology, immunity, and vaccine development. J. Health Popul. Nutr. 18:5-14. [PubMed] [Google Scholar]
- 17.Isegawa, Y., O. Nakagomi, T. Nakagomi, S. Ishida, S. Uesugi, and S. Ueda. 1993. Determination of bovine rotavirus G and P serotypes by polymerase chain reaction. Mol. Cell. Probes 7:277-284. [DOI] [PubMed] [Google Scholar]
- 18.Iturriza-Gómara, M., G. Kang, and J. Gray. 2004. Rotavirus genotyping: keeping up with an evolving population of human rotaviruses. J. Clin. Virol. 31:259-265. [DOI] [PubMed] [Google Scholar]
- 19.Jiang, B., J. R. Gentsch, and R. I. Glass. 2002. The role of serum antibodies in the protection against rotavirus disease: an overview. Clin. Infect. Dis. 34:1351-1361. [DOI] [PubMed] [Google Scholar]
- 20.Kapikian, A. Z., Y. Hoshino, and R. M. Chanock. 2001. Rotaviruses, p. 1787-1825. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott, Williams & Wilkins, Philadelphia, Pa.
- 21.Kapikian, A. Z., L. Simonsen, T. Vesikari, Y. Hoshino, D. M. Morens, R. M. Chanock, J. R. La Montagne, and B. R. Murphy. A hexavalent human-bovine (UK) rotavirus reassortant vaccine designed for use in developing countries and delivered in a schedule with potential to eliminate the risk of intussusception. J. Infect. Dis., in press. [DOI] [PubMed]
- 22.Koshimura, Y., T. Nakagomi, and O. Nakagomi. 2000. The relative frequencies of G serotypes of rotavirus recovered from hospitalized children with diarrhea: a 10-year survey (1987-1996) in Japan with a review of globally collected data. Microbiol. Immunol. 44:499-510. [DOI] [PubMed] [Google Scholar]
- 23.Laird, A. R., J. R. Gentsch, T. Nakagomi, O. Nakagomi, and R. I. Glass. 2003. Characterization of serotype G9 rotavirus strains isolated in the United States and India from 1993 to 2001. J. Clin. Microbiol. 41:3100-3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leite, J. P. G., A. A. Alfieri, P. A. Woods, R. I. Glass, and J. R. Gentsch. 1996. Rotavirus G and P types circulating in Brazil: characterization by RT-PCR, probe hybridization, and sequence analysis. Arch. Virol. 141:2365-2374. [DOI] [PubMed] [Google Scholar]
- 25.Liprandi, F., M. Gerder, Z. Bastidas, J. A. Lopez, F. H. Pujol, J. E. Ludert, D. B. Joelsson, and M. Ciarlet. 2003. A novel type of VP4 carried by a porcine rotavirus strain. Virology 315:373-380. [DOI] [PubMed] [Google Scholar]
- 26.Martella, V., M. Ciarlet, A. Camarda, A. Pratelli, M. Tempesta, G. Greco, A. Cavalli, G. Elia, N. Decaro, V. Terio, G. Bozzo, M. Camero, and C. Buonavoglia. 2003. Molecular characterization of the VP4, VP6, VP7, and NSP4 genes of lapine rotaviruses identified in Italy: emergence of a novel VP4 genotype. Virology 314:358-370. [DOI] [PubMed] [Google Scholar]
- 27.Martella, V., V. Terio, S. Arista, G. Elia, M. Corrente, A. Madio, A. Pratelli, M. Tempesta, A. Cirani, and C. Buonavoglia. 2004. Nucleotide variation in the VP7 gene affects PCR genotyping of G9 rotaviruses identified in Italy. J. Med. Virol. 72:143-148. [DOI] [PubMed] [Google Scholar]
- 28.Mascarenhas, J. D. P., A. C. Linhares, Y. B. Gabbay, and J. P. G. Leite. 2002. Detection and characterization of rotavirus G and P types from children participating in a rotavirus vaccine trial in Belém, Brazil. Mem. Inst. Oswaldo Cruz 97:113-117. [DOI] [PubMed] [Google Scholar]
- 29.McNeal, M. M., K. Sestak, A. H.-C. Choi, M. Basu, M. J. Cole, P. P. Aye, R. P. Bohm, and R. L. Ward 2005. Development of a rotavirus-shedding model in rhesus macaques, using a homologous wild-type rotavirus of a new P genotype. J. Virol. 79:944-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parashar, U. D., E. G. Hummelman, J. S. Bresee, M. A. Miller, and R. I. Glass. 2003. Global illness and deaths caused by rotavirus disease in children. Emerg. Infect. Dis. 9:565-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rahman, M., J. Matthijnssens, T. Goegebuer, K. De Leener, L. Vanderwegen, I. van der Donck, L. van Hoovels, S. de Vos, T. Azim, and M. van Ranst. 2004. Predominance of rotavirus G9 genotype in children hospitalized for rotavirus gastroenteritis in Belgium during 1999-2003. J. Clin. Virol. 33:1-6. [DOI] [PubMed] [Google Scholar]
- 32.Ramachandran, M., C. D. Kirkwood, L. Unicomb, N. A. Cunliffe, R. L. Ward, M. K. Bhan, H. F. Clark, R. I. Glass, and J. R. Gentsch. 2000. Molecular characterization of serotype G9 rotavirus strains from a global collection. Virology 278:436-444. [DOI] [PubMed] [Google Scholar]
- 33.Rao, C. D., K. Gowda, and B. S. Y. Reddy. 2000. Sequence analysis of VP4 and VP7 genes of nontypeable strains identifies a new pair of outer capsid proteins representing novel P and G genotypes in bovine rotaviruses. Virology 276:104-113. [DOI] [PubMed] [Google Scholar]
- 34.Rosa e Silva, M. L., I. P. de Carvalho, and V. Gouvea. 2002. 1998-1999 rotavirus seasons in Juiz de Fora, Minas Gerais, Brazil: detection of an unusual G3P[4] epidemic strain. J. Clin. Microbiol. 40:2837-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rubilar-Abreu, E., K.-O. Hedlund, L. Svenssson, and C. Mittelholzer. 2005. Serotype G9 rotavirus infections in adults in Sweden. J. Clin. Microbiol. 43:1374-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santos, N., C. C. Soares, E. M. Volotão, M. C. M. Albuquerque, and Y. Hoshino. 2003. Surveillance of rotavirus strains in Rio de Janeiro, Brazil, from 1997 to 1999. J. Clin. Microbiol. 41:3399-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santos, N., E. M. Volotao, C. C. Soares, M. C. M. Albuquerque, F. M. da Silva, V. Chizhikov, and Y. Hoshino. 2003. VP7 gene polyorphism of serotype G9 rotavirus strains and its impact on G genotype determination by PCR. Virus Res. 93:127-138. [DOI] [PubMed] [Google Scholar]
- 38.Santos, N., and Y. Hoshino. 2005. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev. Med. Virol. 15:29-56. [DOI] [PubMed] [Google Scholar]
- 39.Souza, M. B. L. D., M. L. Racz, J. P. G. Leite, C. M. Soares, R. M. Martins, V. Munford, and D. D. Cardoso. 2003. Molecular and serological characterization of group A rotavirus isolates obtained from hospitalized children in Goiania, Brazil, 1998-2000. Eur. J. Microbiol. Infect. Dis. 22:441-443. [DOI] [PubMed] [Google Scholar]
- 40.Steyer, A., M. Poljsak-Prijatelj, D. Barlic-Maganja, T. Bufon, and J. Marin. 2005. The emergence of rotavirus genotype G9 in hospitalized children in Slovenia. J. Clin. Virol. 33:7-11. [DOI] [PubMed] [Google Scholar]
- 41.Taniguchi, K., F. Wakasugi, Y. Pongsuwanna, T. Urasawa, S. Ukae, S. Chiba, and S. Urasawa. 1992. Identification of human and bovine rotavirus serotypes by polymerase chain reaction. Epidemiol. Infect. 109:303-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tian, Y., O. Tarlow, A. Ballard, U. Desselberger, and M. A. McCrae. 1993. Genomic concatemerization/deletion in rotaviruses: a new mechanism for generating rapid genetic change of potential epidemiological importance. J. Virol. 67:6625-6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Timenetsky, M. C. S. T., N. Santos, and V. Gouvea. 1994. Survey of rotavirus G and P types associated with human gastroenteritis in São Paulo, Brazil, from 1986 to 1992. J. Clin. Microbiol. 32:2622-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Török, T. J., P. E. Kilgore, M. J. Clarke, R. C. Holman, J. S. Bresee, R. I. Glass, and the National Respiratory and Enteric Virus Surveillance System Collaborating Laboratories. 1997. Visualizing geographic and temporal trends in rotavirus activity in the United States, 1991 to 1996. Pediatr. Infect. Dis. J. 16:941-946. [DOI] [PubMed] [Google Scholar]